xGen™ Pre-Hybridization Capture Normalase™ Module
Accelerated pre-hybridization capture library pooling
Automatable normalization technology that eliminates tedious library concentration adjustments while improving library balance. This solution can easily be integrated into standard library preparation and hybridization capture protocols to increase efficiency and reduce costs.
xGen™ NGS—made to streamline.
Ordering
- Save time and increase throughput—uniform sampling with fewer handling steps to generate balanced library representation for pre-hybridization capture library pooling
- Reduce sequencing costs—improve library balancing and obtain predictable read numbers
- Flexible design—compatible with xGen library preparation and hybridization capture workflows to produce more evenly balanced sequence data
- Reduced hands-on time—a simple 45-minute protocol that seamlessly integrates into NGS workflows
- Automation compatible—streamlined for high-throughput applications
Product details
The xGen Pre-Hybridization Capture Normalase Module uses a novel enzymatic normalization technology for generating equimolar library pools and balanced sample representation ahead of hybridization capture. This method eliminates the need for individual library quantification and pooling of variable volumes. Instead, equal volumes are pooled during the Normalase chemistry. The resulting pool has a sample read depth with a coefficient of variation of (CV) ≤ 10%, which is more uniform than library pools generated using qPCR quantification and fluorometric assays.
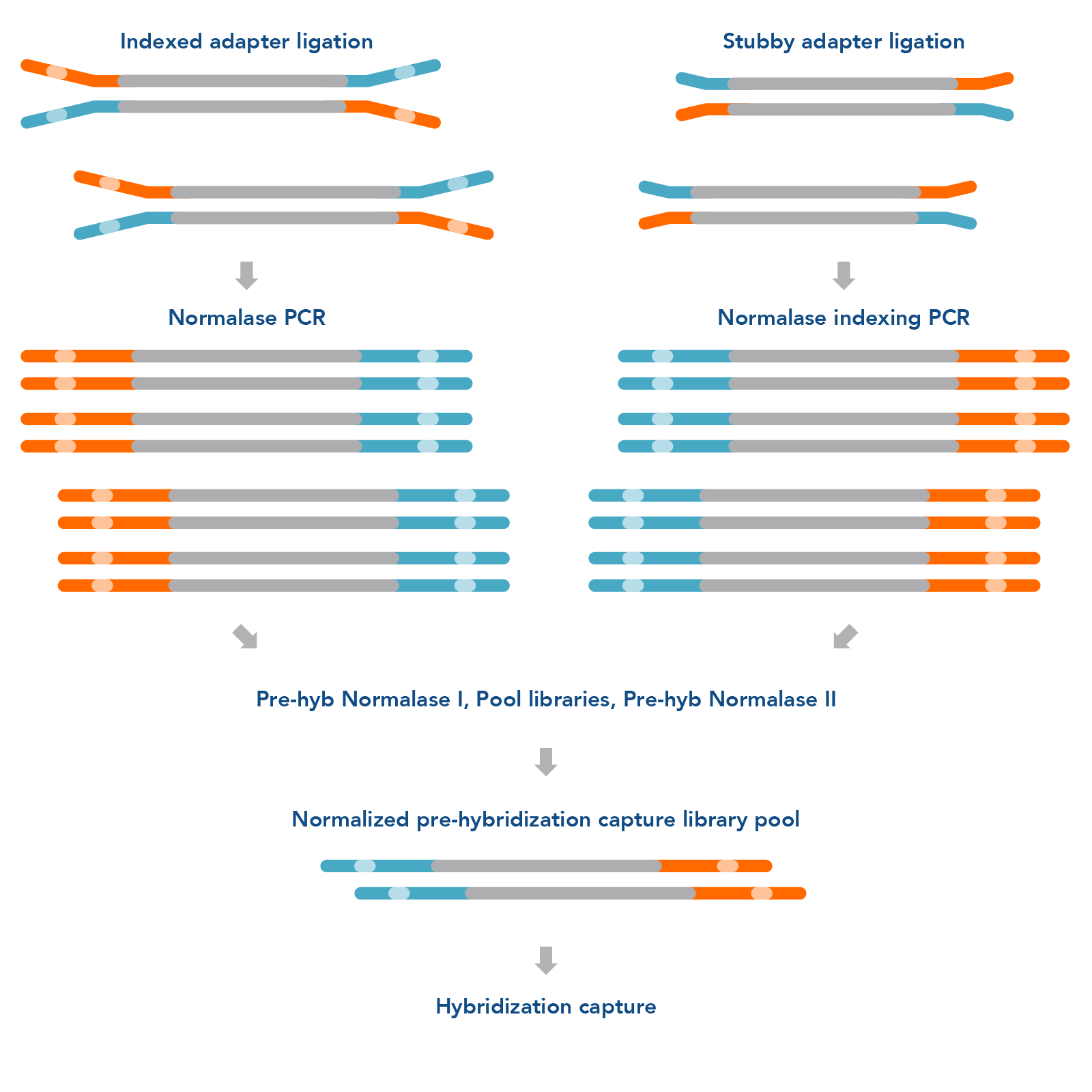
The xGen Pre-Hybridization Capture Normalase Module (Figure 1 ) can be easily integrated into standard DNA and RNA library preparation and hybridization capture protocols to improve turnaround time and loading accuracy for NGS research. The workflow does not require a second PCR; instead, indexing and Normalase library conditioning can occur in the same reaction.
Using the xGen Pre-Hybridization Capture Normalase Module, researchers can select a broad range of library inputs into hybridization capture (100–500 ng), across multiple insert sizes (150–350 bp), while supporting multiplexing of 4–24 libraries per pool.
Figure 1. xGen Pre-Hybridization Capture Normalase Module workflow. The xGen Pre-Hybridization Capture Normalase Module workflow begins after NGS library adapter ligation, using either full-length indexed adapters or truncated adapters, where Normalase PCR primers are used to amplify the libraries above the minimum threshold and condition the libraries for downstream Normalase enzymology. For full-length indexed adapter libraries, xGen Normalase terminal primers are used; for truncated adapter libraries, xGen Normalase Indexing primers are used. Amplified and conditioned NGS libraries are then individually incubated for 15 minutes with the Pre-Hyb Normalase I Master Mix to enzymatically select a specified molarity of each NGS library. After Pre-Hyb Normalase I, each library is pooled using equal volumes into a single tube and incubated for 15 minutes with the Pre-Hyb Normalase II Master Mix which enzymatically normalizes each NGS library to the specified selected molarity. The result is a balanced, multiplexed NGS library pool ready for hybridization capture.
Product data
Fast library normalization that seamlessly integrates into NGS library preparation workflows
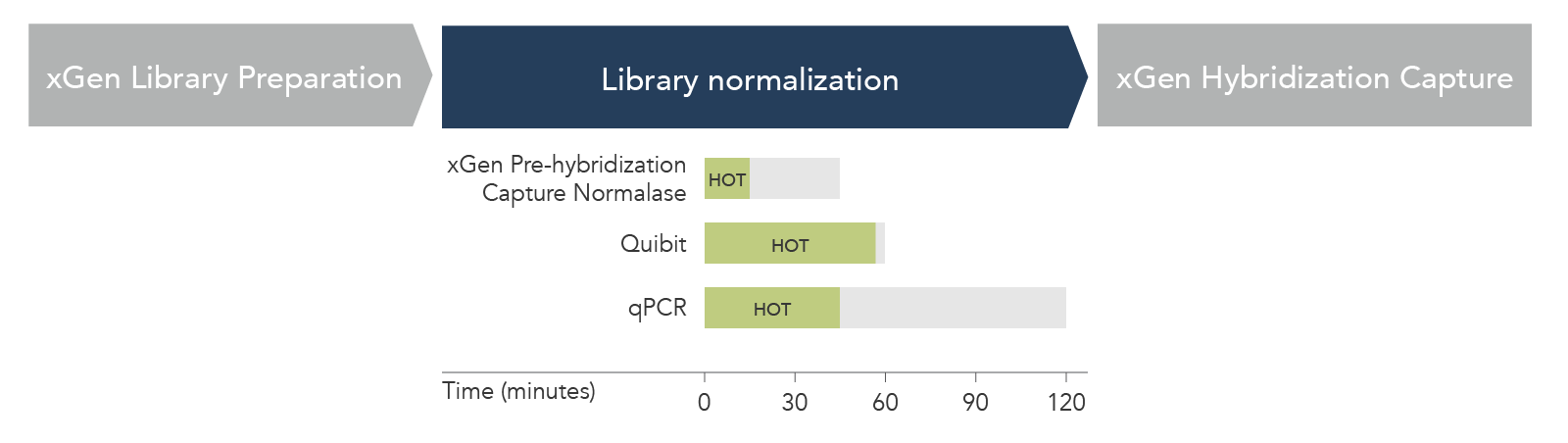
Figure 2. xGen Pre-Hybridization Capture Normalase Module reduces hands-on time (HOT) and total workflow time compared to standard library normalization methods. HOT (green box) and total workflow time (gray box) were compared for n = 24 libraries across xGen Pre-Hybridization Capture Normalase Module, Qubit, and qPCR-based normalization methods. xGen Pre-Hybridization Capture Normalase Module had the shortest HOT (15 min) and total workflow time (45 min).
Better normalization compared to conventional methods
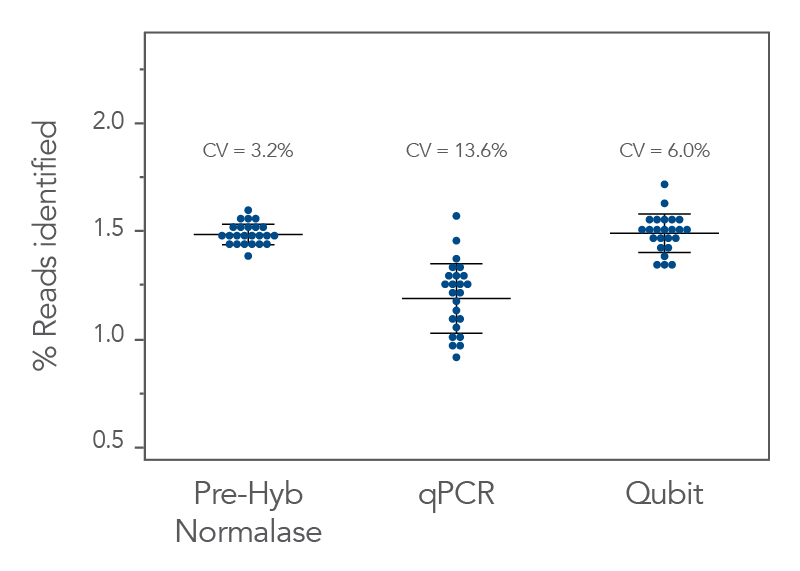
Figure 3. xGen Pre-Hybridization Capture Normalase Module generates better balanced coverage into hybridization capture. xGen DNA EZ libraries (n = 24) were generated with 10 ng of DNA (NA12878) and amplified with xGen Normalase UDI primers using 11 cycles of PCR and normalized to 250 ng for hybridization capture. Libraries were normalized and pooled prior to hybridization capture based on one of the three methods–the xGen Pre-Hybridization Capture Normalase Module workflow, qPCR quantification, or Qubit quantification. Prior to hybridization capture, the libraries were sequenced on a single Illumina® MiniSeq (300 cycle) run, and the coefficient of variation (CV) of the reads measured. CV for the xGen Pre-Hybridization Capture Normalase Module pool was 3.2% showing robust normalization of multiplexed pools compared to standard quantification methods (13.6% for qPCR and 6.0% for Qubit).
Libraries normalized using xGen Pre-Hybridization Capture Normalase Module maintain high on-target rates
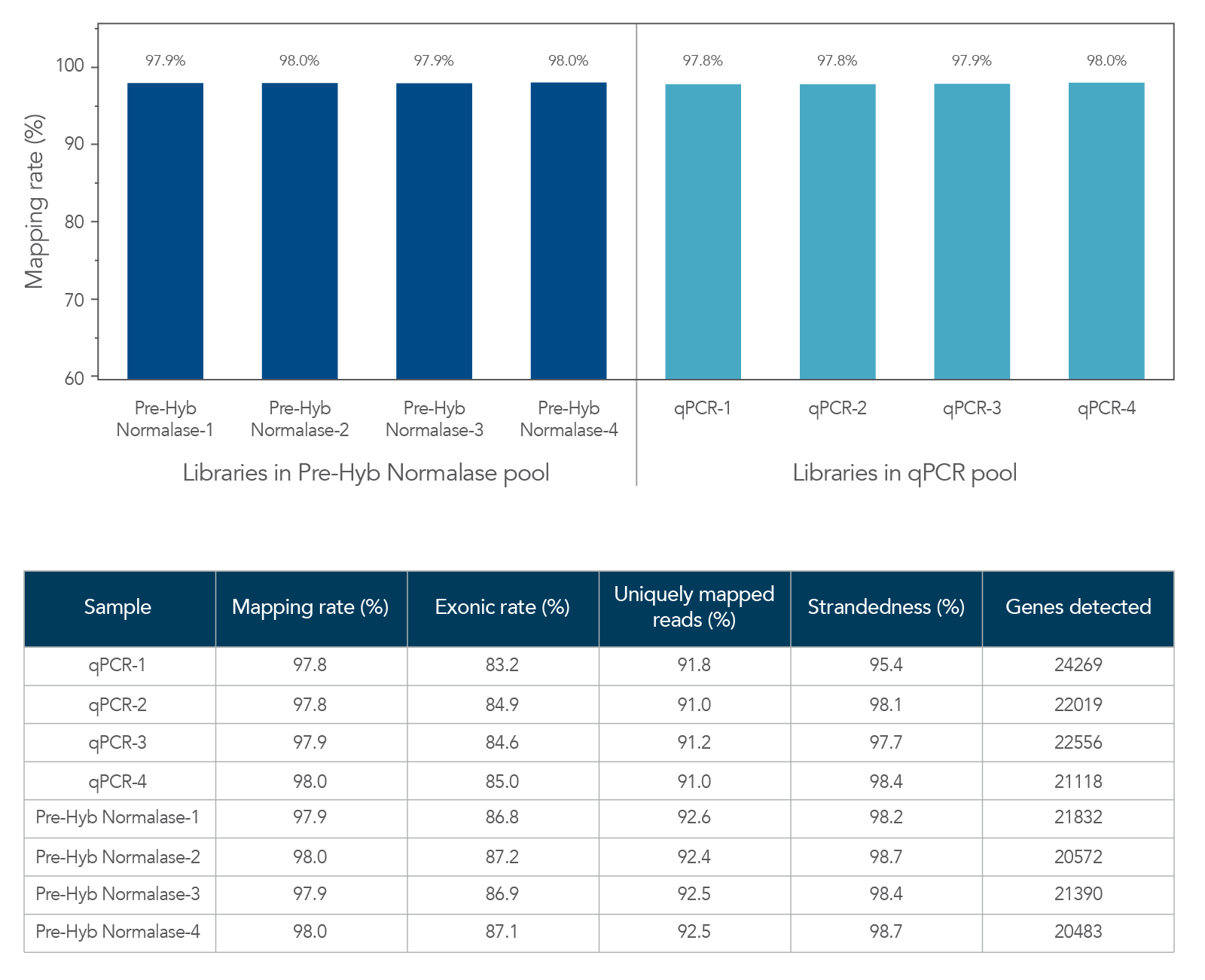
The xGen Pre-Hybridization Capture Normalase Module can be integrated into both DNA and RNA library prep workflows to generate high-quality NGS libraries (Figures 4,5). DNA libraries normalized with the xGen Pre-Hybridization Capture Normalase Module maintained high on-target rates (Figure 4). Similarly, RNA-seq libraries that used the xGen Pre-Hybridization Capture Normalase Module resulted in libraries that had high mapping and exonic rates, as well as high percentages of uniquely mapped reads and strandedness (Figure 5) indicating that the xGen Pre-Hybridization Capture Normalase Module can reduce HOT, while generating well-balanced, high-quality NGS libraries.
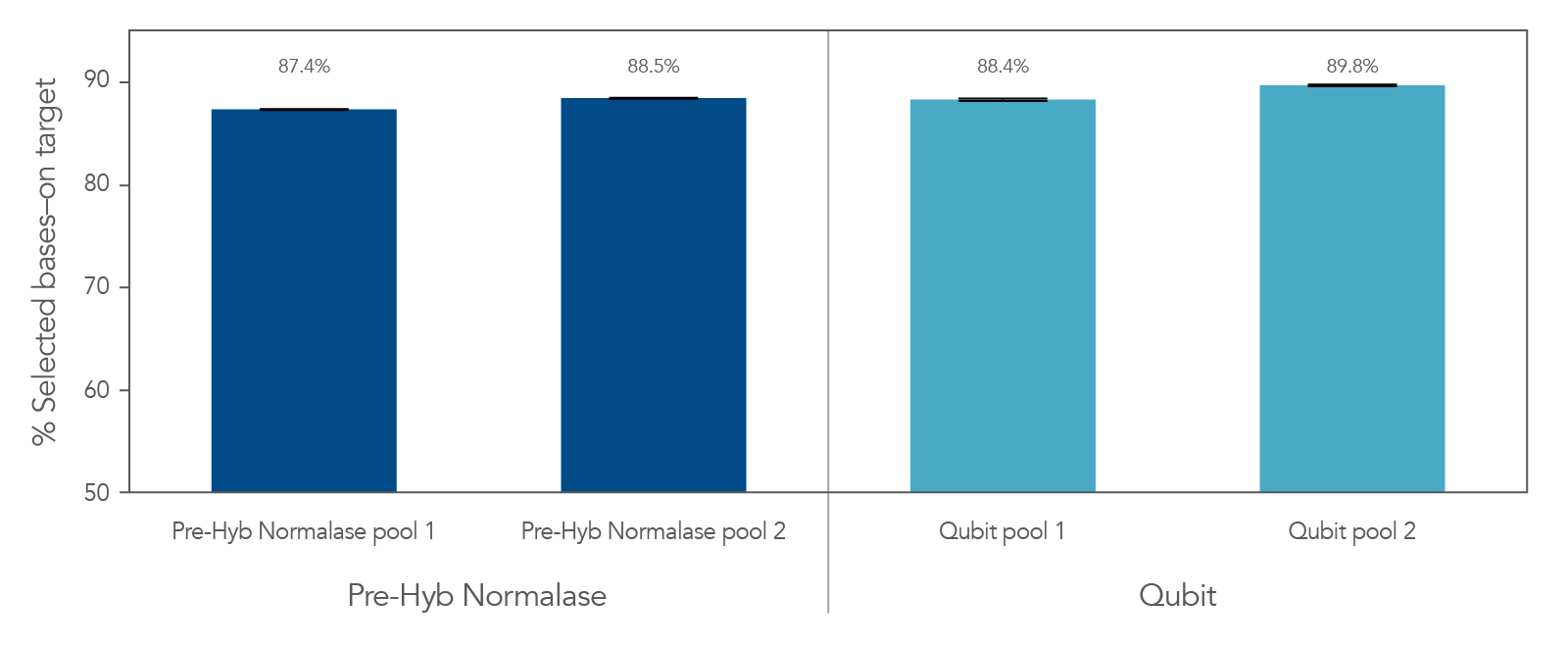
Figure 4. Libraries the integrated the xGen Pre-Hybridization Capture Normalase Module retain similar on-target rates compared to conventional methods. xGen DNA EZ libraries (n = 48) were generated with 10 ng of DNA (NA12878) and amplified with xGen Normalase UDI primers using 11 cycles of PCR. Libraries were separated between 2 normalization methods, Qubit and xGen Pre-Hybridization Capture Normalase Module (n = 12 libraries/pool x 2 pools). Libraries were either normalized and pooled based on Qubit quantification or using equal volume pooling with xGen Pre-Hybridization Capture Normalase Module. 500 ng of these libraries were then enriched with the xGen Exome Hyb Panel v2 and sequenced on the Illumina® NextSeq 2000 system. Error bars represent the standard deviation from the mean for the 12 libraries in each pool.
xGen Pre-hybridization Capture Normalase preserves RNA-seq data quality
Figure 5. Comparable RNA-seq data quality between libraries in xGen Pre-Hybridization Capture Normalase and qPCR pools. xGen Broad Range RNA libraries (n = 4) were generated from 100 ng of UHR total RNA (Universal Human Reference RNA, Thermo Fisher Scientific), amplified, and indexed with xGen Normalase UDI primers using 11 cycles of PCR. Libraries were either normalized and pooled based on qPCR quantification or using equal volume pooling with xGen Pre-Hybridization Capture Normalase Module (n = 4 libraries/normalization method). Library pools were enriched with xGen Exome Hyb Panel v2 and sequenced on an Illumina® NextSeq 2000 P2 flow cell (2 x 149). Sequencing data was normalized to 65 M paired-end reads, mapped using STAR, and analyzed for RNA-seq metrics using Picard.
Resources
Frequently asked questions
I am struggling to achieve the 3X minimum yield with the xGen™ cfDNA & FFPE DNA Library Prep v2 Kit for the maximum input into hybridization capture, how could I proceed with using the xGen™ Pre-Hyb Normalase™ Module 96 rxn?
What kind of samples should I use with xGen™ Pre-Hyb Normalase™ Module 96 rxn?
How much time does it take to run the xGen™ Pre-Hyb Normalization protocol?
How many DNA libraries can I multiplex with xGen™ Pre-Hyb Normalase™ Module 96 rxn?
Which library preparation workflows are supported for the xGen™ Pre-Hyb Normalase™ Module 96 rxn?
The xGen Pre-Hyb Normalase Module 96 rxn is compatible with the following library preparation kits: