xGen™ Broad-Range RNA Library Preparation Kit
Comprehensive data from a wide range of RNA inputs
Compatible with a range of input types and quantities, a variety of indexing options for your research, as well as manual or automated systems. Assemble RNA-Seq libraries from 1st strand cDNA synthesis.
xGen NGS—made for broad range RNA library preparation.
Ordering
- Comprehensive data—for mapping rates, genes identified, and transcript coverage.
- Accommodates your samples with a single kit—consistent results from 10 ng to 1 µg total RNA or 100 pg to 100 ng mRNA input.
- Consistent libraries—minimal adapter dimers so adapter titration is not required for supported inputs.
- Save time, reduce costs—go from RNA to library in 4.5 hours with a wide variety of index options, ranging from up to 1536 UDI primer pairs (available with or without NormalaseTM), and up to 96 CDI primer pairs (with or without Normalase).
- Workflow—designed for easy automation.
Product details
The xGen Broad-Range RNA Library Prep Kit has a fast NGS transcriptomics research workflow with comprehensive transcript coverage and NGS data quality from a broad range of input quantities for Illumina® sequencing platforms. Leveraging proprietary AdaptaseTM technology, this RNA library prep kit enables stranded RNA library construction directly from 1st strand cDNA without the requirement for 2nd strand cDNA synthesis and degradation, or template-switching methods. The kit is compatible with manual and automated workflows, as well as upstream and downstream enrichment and depletion methods. It supports a variety of indexing options in research studies.
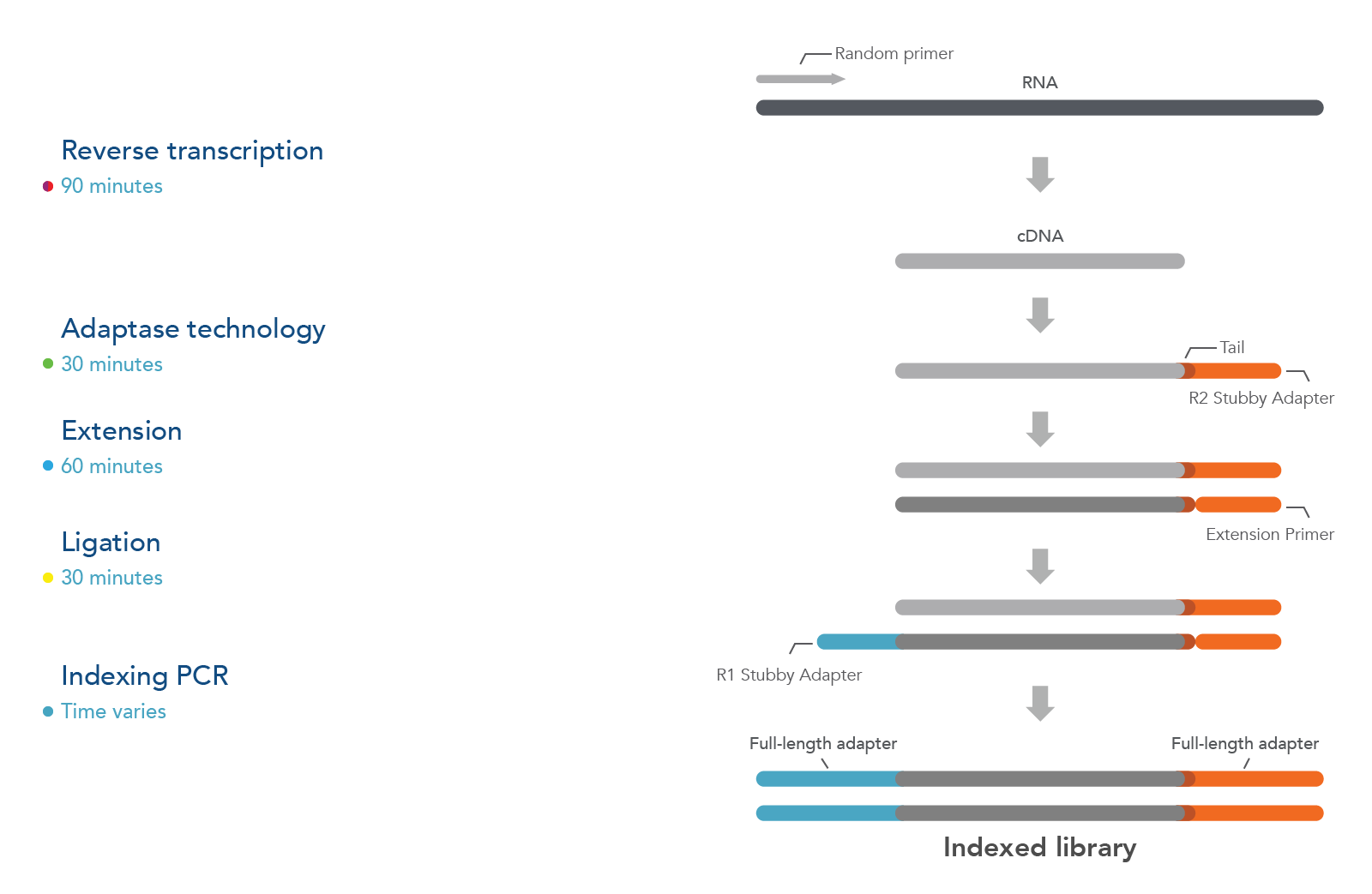
Figure 1. xGen Broad-Range Library Prep workflow: After RNA fragmentation, the reverse transcriptase step uses random primers to generate the first-strand cDNA. Next, Adaptase technology simultaneously performs tailing and ligation to incorporate the R2 Stubby Adapter to the 3’ ends of the cDNA molecules. The extension step produces a dsDNA duplex, while ligation adds the R1 Stubby Adapter to the 3’ ends of the primer-extended cDNA molecules. Finally, indexing PCR increases library yield, incorporates single or dual indexes, and results in full-length adapters at the ends of each molecule. In addition, bead-cleanup steps are needed after extension, ligation, and final indexing PCR steps.
The xGen Broad-Range RNA Library Prep Kit leverages an indexing PCR step to complete the fully indexed adapter sequences, using primers that anneal to the Stubby Adapters, to be fully compatible with Illumina sequencers. IDT supplies a variety of index configurations and strategies, including:
- Combinatorial dual indexing, up to 96 combinations and compatible with Normalase
- Unique dual indexing, up to 1536 unique dual indices and compatible with Normalase
Available in 16, 96, or 4x96-reaction kit sizes, the xGen Broad-Range RNA Library Prep Kit is offered at scales to support evaluation and adoption.
Automation
The xGen Broad-Range RNA Library Prep Kit protocol is readily automatable. A 10% overage volume of reagents is supplied to accommodate automation and additional reagent overage volume is available upon request.
Table 1. xGen Broad-Range RNA Library Prep Kit specifications.
| Feature | Specification | Benefit |
|---|---|---|
| Input quantity | 10 ng to 1 µg total RNA 100 pg to 100 ng mRNA | Supports a wide input range Consistent library output |
| RNA types supported | Poly(A)-enriched mRNA Ribo-depleted RNA Total RNA | Supports most RNA applications |
| Technology | Adaptase tailing and ligation of 1st strand cDNA | No 2nd strand cDNA No adapter titration Fewer dimers and duplicates detected in internal research studies (Figure 2) Maintains strandedness (≥97%) Higher mapping, transcript detection |
| Workflow time* | 4.5 hours | Less hands-on time |
| Kit reaction sizes | 16, 96, and 4x96 | Evaluation and adoption |
| Components provided | Fragmentation module RT module Library prep Polymerase | Complete solution for processing total, enriched, or depleted RNA from transcript to library |
| Indexing options | Combinatorial dual Unique dual Normalase compatible | Flexible for different sequencers, workflows, and applications |
| Multiplexing capability | Up to 1536 libraries | Save sequencing costs |
| Automation | Compatible with liquid handlers Custom packaging available | Supports high-throughput applications |
*Workflow time is based upon incubation times and expected times for hands-on-steps. Actual workflow time may vary depending on individual factors in your laboratory.
Product data
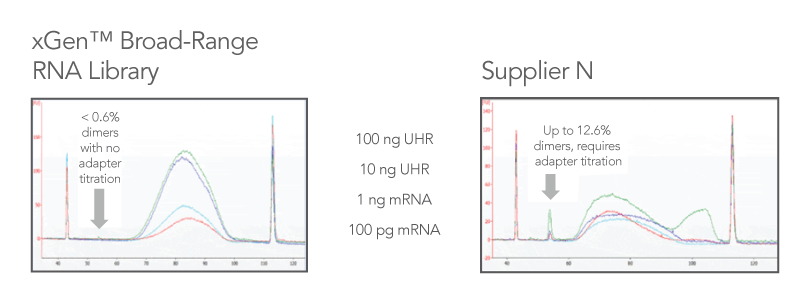
Reduced dimer formation means no adapter titration
Adaptase technology results in minimal adapter dimers, which means no need for adapter titration (Figure 2). Compared to leading RNA library kits, which can produce libraries with >10% adapter dimers and require adapter titration steps, the xGen Broad-Range RNA Library Prep Kit produces <1% adapter dimers and maintains ligation efficiency at all supported input levels.
Figure 2. Comparison of kits. Libraries were prepared using two different kits with the same quantity of input material (n = 1 per input quantity) and subjected to the same number of PCR cycles according to the xGen Broad-Range
protocol recommendations during library amplification. Representative library sizes, yields, and BioanalyzerTM (Agilent) traces illustrate typical libraries generated from the same input series of poly(A)-enriched Universal Human Reference
(UHR) RNA (Agilent 740000) or human brain mRNA (Takara 636102), when processed by either the xGen Broad-Range RNA Library Prep Kit (left panel) or an RNA library kit from an alternate supplier (right panel). The arrows indicate adapter dimers that
were generated during library preparation.
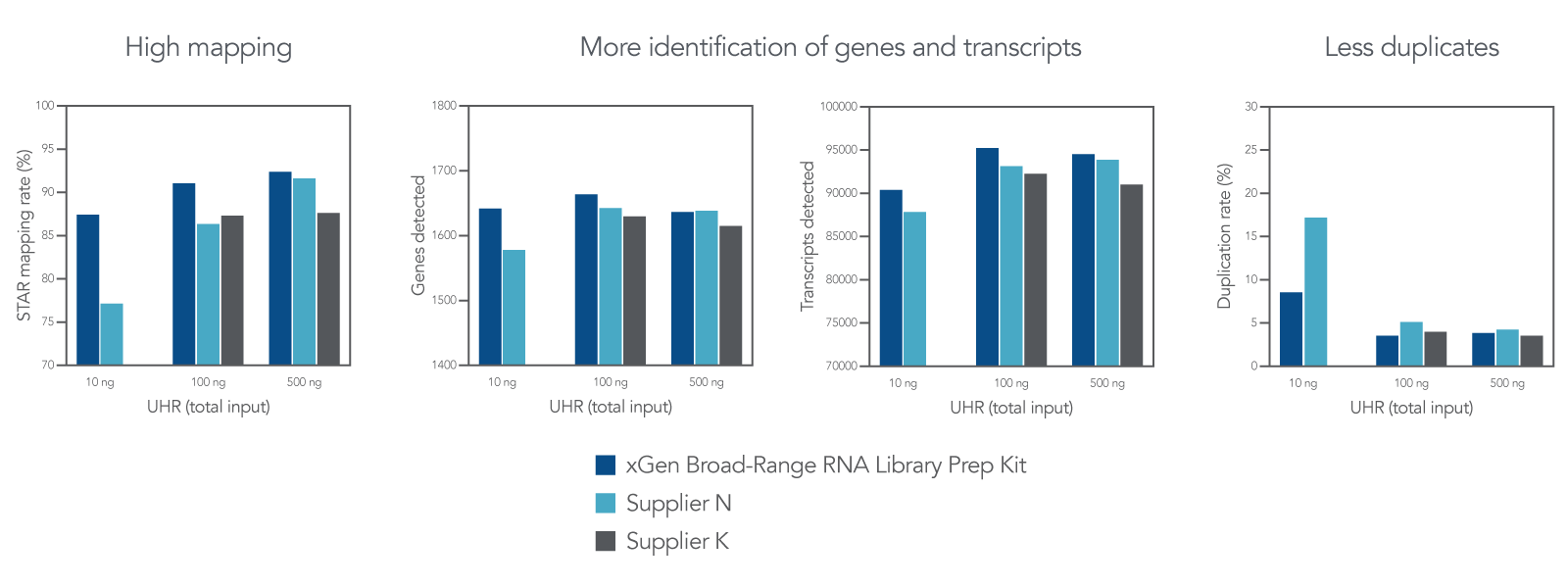
High mapping and transcript identification with lower duplication rates
Figure 3. Comparison of data obtained using different suppliers’ kits. Universal Human Reference (UHR) Total RNA (Agilent 740000) was enriched using the NEBNext® poly(A) mRNA Magnetic Isolation Module (NEB E7490), before being processed by the xGen Broad-Range RNA Library Prep Kit and other supplier kits K and N. For each kit evaluated, libraries were prepared at 10, 100 and 500 ng input, where n = 1 sample per input quantity. PCR amplification of each library was performed as follows: 10 cycles for 500 ng inputs, 12 cycles for 100 ng inputs, and 16 cycles for 10 ng inputs. Libraries were sequenced on a MiniSeqTM with 2x75 bp paired-end reads. Fastq files were downsampled to 2.2 million reads before analysis using STAR (mapping rate), RNASeqC (genes/transcripts detected), or Picard (duplication rate). The xGen Broad-Range RNA Library Prep Kit has high mapping percentage, detects more genes and transcripts, and has fewer duplicates.
Resources
Frequently asked questions
What are the new names of the Swift products?
The Swift products were rebranded and now belong to the xGen™ NGS product line. The following lists the old Swift product name, and then the new name hyperlinked to its current product page.
| Swift product name | New IDT product name |
|---|---|
| Accel-NGS® Adaptase® Module for Single-Cell Methyl-Seq | xGen Adaptase™ Module |
| Normalase® Amplicon Panels (SNAP) Core | xGen Amplicon Core |
| SARS-CoV-2 Additional Genome Coverage Panel | xGen SARS-CoV-2 Expanded Amplicon Panel |
| SARS-CoV-2 S Gene Panel | xGen SARS-CoV-2 SGene Amplicon Panel |
| SNAP Set 1A Combinatorial Dual Indexing Primers | xGen Amplicon CDI Primers |
| Swift Combinatorial Dual Indexing Primers | xGen CDI Primers |
| Swift Normalase® Combinatorial Dual Indexing Primers | xGen Normalase CDI Primers |
| Accel-NGS 1S Plus DNA Library | xGen ssDNA & Low-input DNA Library Prep |
| Swift 2S Sonic DNA Library | xGen DNA Library Prep MC |
| Swift 2S Sonic Flexible DNA Library | xGen DNA Library Prep MC UNI |
| Swift 2S Turbo v2 | xGen DNA Library Prep EZ |
| Swift 2S Turbo Flexible v2 DNA Library | xGen DNA Library Prep EZ UNI |
| Accel-NGS Methyl-Seq DNA Library | xGen Methyl-Seq Library Prep |
| Swift Normalase | xGen Normalase Module |
| Swift Rapid RNA Library | xGen RNA Library Prep Kit |
| Swift RNA Library | xGen Broad-Range RNA Library Prep |
Does the xGen™ Broad-Range RNA Library Prep Kit include upstream poly(A) enrichment or ribodepletion modules?
No, this kit takes as input one of the following:
- Total RNA
- mRNA that has already been enriched by poly(A) selection
- RNA that has already been depleted of rRNA
The xGen Broad-Range RNA Library Prep Kit begins with an RNA fragmentation and reverse transcription step. Therefore, if mRNA enrichment or ribodepletion is required, see the xGen Broad-Range RNA Library Prep Kit Protocol (Appendix A) for kits that have been tested for compatibility with this protocol.
Also, refer to the protocol for details on matching the output of these upstream modules with the initial steps in the xGen Broad-Range RNA Library Kit instructions.
| Method | Recommended kit | Alternate kit |
|---|---|---|
| Poly(A) mRNA Enrichment | NEBNext® Poly (A) mRNA Magnetic Isolation Module Cat. No. E7490 |
Lexogen Poly (A) RNA Selection Kit Cat. No. 039.100 |
| Ribodepletion | Lexogen RiboCop rRNA Depletion Kit V1.2 (Human/Mouse/Rat) Cat. No. 037.24/96 |
NEBNext rRNA Depletion Kit (Human/Mouse/Rat) Cat. No. E6310S/L/X |
What indexing primers can I use with this xGen™ Broad-Range RNA Library Kit?
The xGen™ Broad-Range RNA Library Prep Kit has an Indexing PCR step to complete the fully indexed adapter sequences following ligation of stubby adapters during the library prep workflows, making it fully compatible with Illumina® sequencers.
IDT supplies a variety of index configurations and strategies, including dual indexing as either:
- Combinatorial dual indexing, up to 96 combinations
- Unique dual indexing, up to 1536 combinations
What are the main steps for library construction using the xGen™ Broad-Range Library Prep Kit?
The xGen™ Broad-Range RNA Library Prep Kit has 5 main steps:
- Fragmentation & Reverse Transcription, where RNA is fragmented and converted to 1st strand cDNA
- Adaptase, where the Read 2 Stubby Adapter is added to the 3’ end of 1st strand cDNA
- Extension, where a primer extension reaction generates a dsDNA duplex
- Ligation, where the Read 1 Stubby Adapter is ligated to the duplex
- Indexing PCR, where the fully indexed adapter sequences are added, and the library is amplified to generate sufficient yield for sequencing.
What are the input requirements for xGen™ Broad Range RNA Library Prep Kit?
How do I quantify and characterize xGen™ Broad-Range RNA libraries?
There are many commercially available kits suitable for library quantification. Following the recommended PCR cycles in the xGen Broad-Range RNA Library Prep Kit Protocol will result in a library concentration of at least 4 nM.
The xGen Broad-Range RNA Library Prep Kit can also be used with the Normalase™ Module to generate 2 nM or 4nM normalized library pools.
Is there a recommended bioinformatics workflow for the xGen™ Broad-Range Library Prep Kit?
Adaptase™ technology, used in the xGen™ Broad-Range RNA Library Prep Kit, adds a low complexity polynucleotide tail with a median length of eight bases to the 3’ end of each fragment during the addition of the first NGS adapter molecule. Considering this, it is normal and expected to observe them at the beginning of Read 2 (R2).
When read length is close to fragment size, the tail may also be observed toward the end of Read 1 (R1). We recommend using the STAR aligner (Dobin et al. 2013) to soft clip the synthetic tail sequence and provide efficient mapping without extra processing of your sequencing data.
For more details, you can refer to the Appendix section of the xGen Broad-Range RNA Library Prep Kit Protocol pertaining to Data Analysis and Informatics.
Does the xGen™ Broad-Range RNA Library Prep Kit work with damaged or degraded RNA?
Yes.
Refer to the xGen Broad-Range RNA Library Prep Kit Protocol (Appendix B) for details regarding the changes to RNA fragmentation time and appropriate SPRI bead ratios for samples with RIN scores lower than 7.
Note: We do not recommend generating libraries from starting RNA with an RIN score < 2.
What is the shelf life and recommended storage conditions for the xGen™ Broad-Range RNA Library Prep Kit?
The shelf life of the xGen Broad-Range RNA Library Prep Kit is at least 6 months from the time the product is delivered, when stored as required at –20°C and handled according to the protocol.
The enzymes provided in this kit are temperature sensitive, and appropriate care should be taken during handling and storage. Upon receipt, store the kits at –20°C.Is titration of adapters required for lower inputs with the xGen™ Broad-Range RNA Library Prep Kit?
No.
The xGen™ Broad-Range RNA Library Prep Kit does not require adapter titration at any of the supported input levels. Adapter ligation efficiency is maintained at all supported input quantities.
Is the xGen™ Broad-Range RNA Library Prep Kit compatible with my own indexing primers?
See the Appendix section of the xGen Broad-Range RNA Library Prep Kit Protocol pertaining to Indexed Adapter Sequences for information about the adapter sequences and structure of the xGen Broad-Range RNA Library adapters and indexing primer kits.
Contact Scientific Application Support at applicationsupport@idtdna.com if you wish to confirm compatibility of your own custom indexing primers with the stubby adapters supplied in the xGen Broad-Range RNA Library Prep Kit workflows.
Can I generate RNA libraries of different size ranges using the xGen™ Broad-Range RNA Library Prep Kit?
Yes.
See the Appendix section of the xGen™ Broad Range RNA Library Prep Kit Protocol about expected results with alternate fragmentation times.
You can also refer to Appendix E for details on fragmentation times and bead cleanup ratios required for different intended mean library sizes ranging from 380−480 bp (mean insert sizes ranging from 250−350 bp).