xGen™ ssDNA & Low-Input DNA Library Preparation Kit
Rescue valuable data from degraded samples
The xGen ssDNA & Low-Input DNA Library Prep Kit enables preparation of libraries from degraded and damaged ssDNA and dsDNA samples in a single reaction. This kit allows users to sequence difficult-to-process samples via unique, proprietary AdaptaseTM technology.
xGen NGS—made for low-input DNA library preparation.
Ordering
- Low input, high complexity—inputs as low as 10 pg from highly degraded samples
- Compatible with ssDNA and dsDNA samples—preserves input fragmentation patterns for precise mapping of DNA insert ends
- Minimize sequence bias from PCR— high-fidelity PCR polymerase, unique chemistry adapts short DNA fragments ≥40 bp long
- Multiplex up to 1536 samples—pre-plated UDI primers enable multiplexing of up to 1536 samples
- Compatible with xGen NormalaseTM technology—enzymatic normalization of libraries from a single pool of libraries
- Simple 2-hour protocol
- Workflow design for easy automation
Product details
NGS DNA library construction from samples with damaged DNA
The xGen ssDNA & Low-Input DNA Library Prep Kit enables library preparation from damaged samples that can be difficult to obtain sequence information. The xGen ssDNA & Low-Input DNA Library Prep Kit can create libraries in 2 hours from as little as 10 pg of input material. Libraries can be prepared from heavily damaged samples by using IDT Adaptase technology. Unlike other library methods, the Adaptase technology generates library molecules from single-stranded DNA fragments, which allows better recovery of sample input DNA complexity from heavily nicked and degraded samples compared to other commercially available products for Illumina® platforms. Sample types include degraded FFPE, ancient DNA, chromatin immunoprecipitation (ChIP), and other enriched samples that have undergone DNA damage.
Rescue valuable sequencing data from low-abundance samples with ssDNA content
The xGen ssDNA & Low-Input DNA Library Prep Kit is designed for users needing to sequence samples containing ssDNA for research. These include metagenomic and viromic samples where relative abundance of both ssDNA and dsDNA phage and viral genomes can be identified. Other compatible sample types include heat-denatured pathogenic samples, samples extracted under harsh denaturing conditions to decrease extraction bias, and oncology-related liquid biopsy samples when detection of single-stranded cfDNA content is desired.
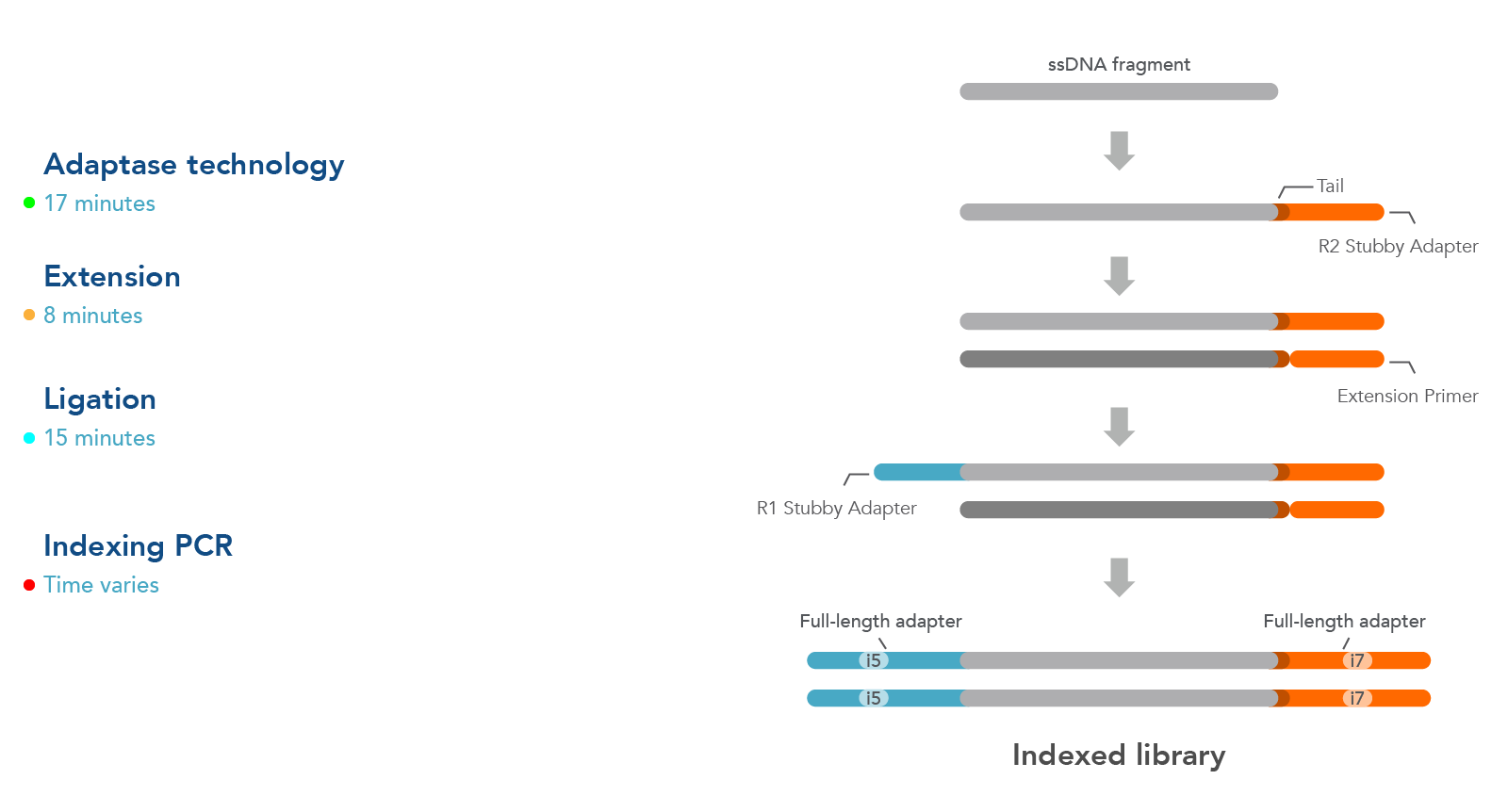
Workflow
The workflow for this kit is shown in Figure 1. As illustrated in this diagram, samples undergo four steps:
- Adaptase technology—the Adaptase reaction simultaneously performs tailing and ligation of R2 Stubby Adapter to the 3´ ends in a highly efficient, template-independent manner.
- Extension—the extension reaction generates a second strand prior to ligation.
- Ligation—ligation reaction adds R1 Stubby Adapter to the original strand.
- Indexing PCR—PCR is used to incorporate sample indexes and sequences needed for Illumina® sequencing. Indexing primers are supplied separately.
Figure 1. Workflow used for the xGen ssDNA & Low-Input DNA Library Prep Kit. Four steps are followed as described in the text.
Specifications
| Feature | xGen ssDNA & Low-Input DNA Library Prep Kit |
|---|---|
| Sample types | Low-quality degraded DNA, ssDNA, mixtures of ssDNA and dsDNA |
| Input amount | 10 pg to 250 ng |
| Indexing compatibility | CDI primers up to 96-plex UDI primers up to 1536-plex |
| System compatibility and multiplexing format | Illumina® sequencing instruments |
| Workflow compatibility | Manual & automated. For list of liquid handlers and scripts, please inquire. |
Product data
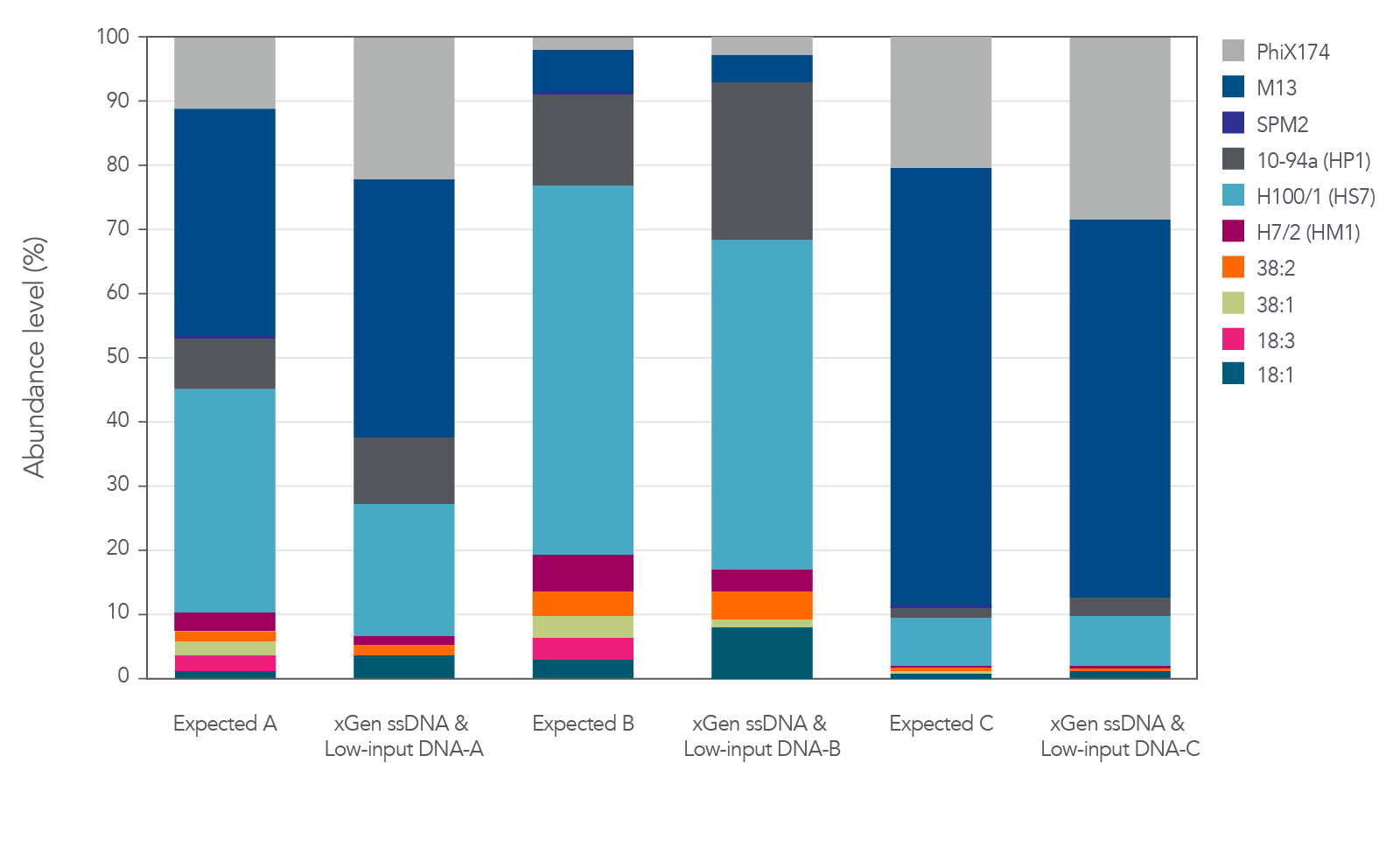
Relative abundance of both ssDNA and dsDNA phage
The xGen ssDNA & Low-Input DNA Library Prep Kit was used to prepare and sequence three artificial viromes containing different proportions of the ssDNA phage PhiX174 and M13 mixed with dsDNA phage (Figure 2). In this experiment, all cases supported
that the proportions were preserved when sequenced with the xGen ssDNA & Low-Input DNA Library Prep Kit without any prior whole genome amplification for identification of ssDNA phage.
The libraries were sequenced on an Illumina® MiSeq® using v2 chemistry and 101 paired-end reads. Reads were then aligned with bowtie2 to assess the relative abundance of each virus, defined as the number of reads mapped normalized
by genome size. These virome abundance values were then compared to the copy number of each genome in the AVC, estimated through SYBR® Green counts for the dsDNA phages, or calculated based on the quantity of DNA measured with NanoDrop® (Thermo Fisher) readings for ssDNA phages. The expected abundance of dsDNA phages was doubled to correct for the presence of two DNA molecules that can act as potential templates compared to a single potential template for ssDNA viruses.
Figure 2. Libraries developed from mixtures of ssDNA and dsDNA phage. Three artificial viral communities (AVCs) were prepared by mixing the isolated gDNA from two ssDNA phages (M13 and PhiX174) with gDNA isolated from
eight dsDNA phages (SPM2, 10-94a, H100/1, H7/2, 38:2, 38:1, 18:3, and 18:1) at different ratios of ssDNA to dsDNA. Artificial viral community A contained 50% ssDNA and 50% dsDNA phage, AVC B contained 10% ssDNA and 90% dsDNA phage, and AVC C contained
90% ssDNA and 10% dsDNA phage. The AVCs were sheared to 200 bp using an M220 Covaris® instrument according to the manufacturer’s instructions, and NGS libraries were constructed utilizing the xGen ssDNA & Low-Input DNA Library Prep Kit.
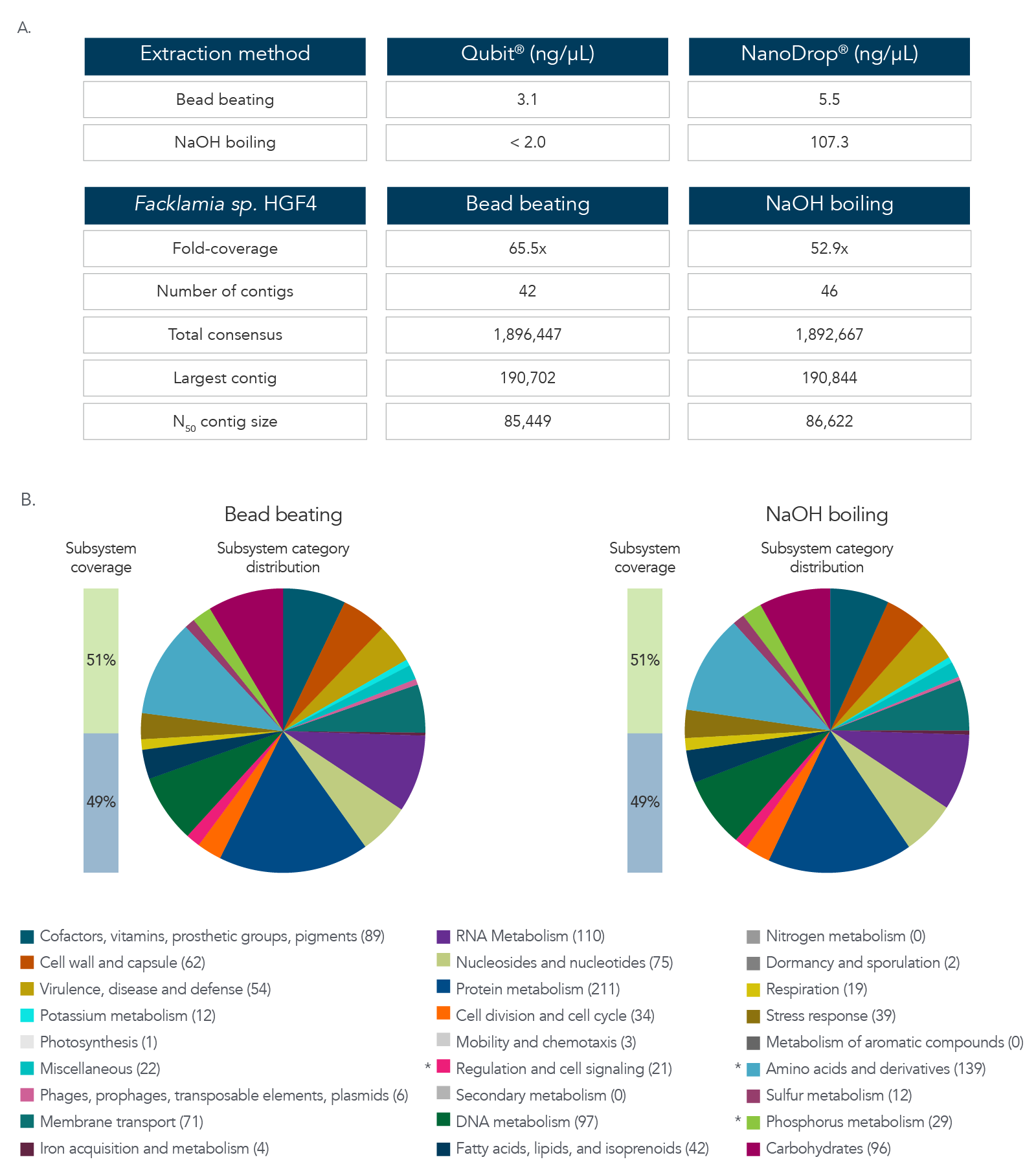
DNA extraction and sequencing of a hard-to-extract microbe
The data in Figure 3A and B show results after DNA extraction and sequencing of Facklamia sp., a microbe recognized for the difficulty it presents for DNA extraction. Pure cultures of Facklamia sp. HGF4 were grown up to 2 x 109 CFU/mL. Facklamia sp. HGF4 was selected because it is a Gram-positive isolate with gDNA that is difficult to extract using a nondenaturing bead-beating extraction method. Because Facklamia sp. HGF4 does not have a reference genome, de novo assembly of the sequencing output was required. Bacterial culture was split into two aliquots—one from which gDNA was isolated using bead beating, and one from which the gDNA was extracted by denaturing NaOH boiling. Briefly, 1 mL of 2 x 109 CFU/ml bacteria was pelleted by centrifugation, the supernatant was removed, and cells were resuspended in 50 µL of sterile water. Then, 50 µL of 0.1 N NaOH was added to the bacterial suspension, and the sample was mixed by vortexing and then boiled at 95 °C for 15 minutes. The solution was then chilled on ice and neutralized by adding 8 µL of Tris-HCl, pH 7.0. The sample was centrifuged at 15,000 x g for two minutes to pellet the cell debris and any unlysed cells, and the supernatant was transferred to a new reaction tube. The isolated gDNA was precipitated with isopropanol to clean and concentrate the sample. DNA recovery was quantified by Qubit® (Thermo Fisher) for dsDNA and NanoDrop for ssDNA. Libraries were prepared using the xGen ssDNA & Low-Input DNA Library Prep Kit. Facklamia sp. HGF4 was assembled de novo using MIRA 4.0 and annotated using the RAST server.
Figure 3. DNA extraction and sequencing of a hard-to-extract microbe. (A) DNA extraction by NaOH boiling produced higher DNA yields from Facklamia sp. than bead beating, and in less time. (B)
Sequencing of the NaOH-extracted DNA produced a comprehensive, de novo assembled genome sequence that was indistinguishable from that produced from bead-beating extracted DNA.
Resources
Frequently asked questions
Can the xGen™ ssDNA & Low-input Library Prep Kit be used with ancient DNA?
Yes. This technology can convert the short, single-stranded fragments,
common in ancient DNA, into NGS library molecules.
However, xGen ssDNA & Low-Input Library Prep Kit is not compatible with ancient DNA samples that are known to have uracil-containing DNA. Instead, use the xGen Methyl-Seq DNA Library Prep Kit which is compatible with uracil-containing DNA.
What are the new names of the Swift products?
The Swift products were rebranded and now belong to the xGen™ NGS product line. The following lists the old Swift product name, and then the new name hyperlinked to its current product page.
| Swift product name | New IDT product name |
|---|---|
| Accel-NGS® Adaptase® Module for Single-Cell Methyl-Seq | xGen Adaptase™ Module |
| Normalase® Amplicon Panels (SNAP) Core | xGen Amplicon Core |
| SARS-CoV-2 Additional Genome Coverage Panel | xGen SARS-CoV-2 Expanded Amplicon Panel |
| SARS-CoV-2 S Gene Panel | xGen SARS-CoV-2 SGene Amplicon Panel |
| SNAP Set 1A Combinatorial Dual Indexing Primers | xGen Amplicon CDI Primers |
| Swift Combinatorial Dual Indexing Primers | xGen CDI Primers |
| Swift Normalase® Combinatorial Dual Indexing Primers | xGen Normalase CDI Primers |
| Accel-NGS 1S Plus DNA Library | xGen ssDNA & Low-input DNA Library Prep |
| Swift 2S Sonic DNA Library | xGen DNA Library Prep MC |
| Swift 2S Sonic Flexible DNA Library | xGen DNA Library Prep MC UNI |
| Swift 2S Turbo v2 | xGen DNA Library Prep EZ |
| Swift 2S Turbo Flexible v2 DNA Library | xGen DNA Library Prep EZ UNI |
| Accel-NGS Methyl-Seq DNA Library | xGen Methyl-Seq Library Prep |
| Swift Normalase | xGen Normalase Module |
| Swift Rapid RNA Library | xGen RNA Library Prep Kit |
| Swift RNA Library | xGen Broad-Range RNA Library Prep |
What is the new name of the xGen™ Lotus™ DNA Library Prep Kit?
IDT has expanded its standard DNA NGS library prep kit offerings to include:
- xGen DNA Library Prep Kit MC compatible with mechanical shearing
- xGen DNA Library Prep Kit EZ that includes the reagents for enzymatic fragmentation
Another alternative is the:
- xGen ssDNA & Low-Input DNA Library Prep Kit for degraded DNA samples (formerly known as Accel-NGS® 1S Plus DNA Library)
Does the Adaptase™ tail need to be bioinformatically trimmed for methylation sequencing?
Yes.
Bases added to the 3’ termini of DNA fragments during the Adaptase reaction contain unmethylated cytosines, which adds both artifactual sequence and methylation information to the dataset. Therefore, tail trimming of xGen™ Methylation-Sequencing DNA libraries is required to attain sufficient mapping efficiency and precise methylation information.
See our technical note, Tail Trimming for Better Data for further details on the tail and how to trim it.
Should the xGen™ ssDNA & Low-input DNA Library Prep Kit be used for FFPE samples?
In most cases using FFPE samples, we would recommend using the xGen cfDNA & FFPE DNA Library Prep Kit.
In scenarios with heavily nicked or denatured DNA resulting from decrosslinking or other high temperature steps, the xGen ssDNA & Low-Input DNA Library Prep Kit can yield higher recovery and conversion rates of input DNA.