Ultramer™ DNA Oligonucleotides
Long, high-quality oligos for demanding applications such as cloning, ddRNAi, homology-directed repair, and gene construction
Ultramer DNA Oligonucleotides are generated by proprietary synthesis methods that deliver high-quality oligos up to 200 bases. They are available single- or double‑stranded and can be delivered in tubes or plates.
Ordering
- Available single- or double-stranded and can be delivered in tubes or plates
- Unrivalled control of oligo specifications with custom formulation and mixing options
Looking for a CRISPR donor? Try our Alt-R™ HDR donor oligos, ideal for point mutations and shorter inserts, or our Alt-R HDR donor blocks for inserts >120 bp. Both are modified and optimized to increase your HDR rates.
Ultramer DNA Oligos
Formulated to 4 nmol or 20 nmol guaranteed yield. Shipped dry or resuspended to 100 µM in IDTE buffer, pH 8.0.
Duplexed Ultramer DNA Oligos
2 oligos, annealed and delivered in a single tube. Formulated to 4 nmol or 20 nmol guaranteed yield. Shipped dry.
The following annealing fees will be applied to each duplex ordered:
Ultramer DNA Oligos
Shipped dry or resuspended to your specifications. Minimum 24 and 96 oligos required for 96- and 384-well plates, respectively. 200 pmol scale requires a minimum of 96 oligos.
Duplexed Ultramer DNA Oligos
1 oligo pair per well, annealed. Shipped dry or resuspended in duplex buffer. Minimum of 24 and 96 oligos required for 96- and 384-well plates, respectively.
For ordering inquiries, please contact us.
Product details
Ultramer DNA Oligos are long, single- and double-stranded synthetic DNA sequences. They are manufactured with a synthesis platform that consistently results in coupling efficiencies over 99.6%. Ultramer DNA Oligos can be used in a variety of molecular biology applications, including (but not limited to):
- Site directed mutagenesis—used to efficiently and quickly generate large insertions, deletions, or change stretches of sequence identity
- In vitro transcription—templates for the synthesis of RNA
- DNA-directed RNAi (ddRNAi)—hairpins are cloned into an expression vector, such as the GeneClip™ U1 Hairpin Cloning System (Promega)
- qPCR—as DNA standards
- CRISPR genome editing—as HDR templates
Every Ultramer DNA Oligo you receive will be deprotected and desalted to remove small molecule impurities. In addition, your oligos will be checked via electrospray ionization mass spectrometry (ESI-MS)* and quantitated by UV spectrophotometry.
*With the exception of mixed base oligos, which could potentially represent multiple sequences and therefore cannot be evaluated by ESI‑MS.
Available modifications for Ultramer DNA Oligos
| Description | 200 pmol Ultramer DNA Oligo | 4 nmol Ultramer DNA Oligo | 20 nmol Ultramer DNA Oligo | PAGE Ultramer DNA Oligo |
|---|---|---|---|---|
| Availability | Plates | Plates and tubes | Plates and tubes | Tubes |
| 5' Phosphorylation | • | • | • | • |
| 5' Biotin | • | • | • | • |
| 5' Amino Modifier C6 | • | • | • | |
| 5' Amino Modifier C12 | • | • | • | |
| 5' deoxyInosine | • | • | • | • |
| 5' deoxyUridine | • | • | • | • |
| Int deoxyInosine | • | • | • | • |
| Int deoxyUridine | • | • | • | • |
| Phosphorothioate Bond | • | • | • | • |
| 2' O-Methyl RNA bases | • | • | • | • |
| 3' Amino Modifier | • | • | ||
| 5' 5-Methyl dC | • | • | • | |
| Int 5-Methyl dC | • | • | • | |
| Int Spacer 18 | • | • | • | |
| 3' C3 Spacer | • | • | • |
Product data
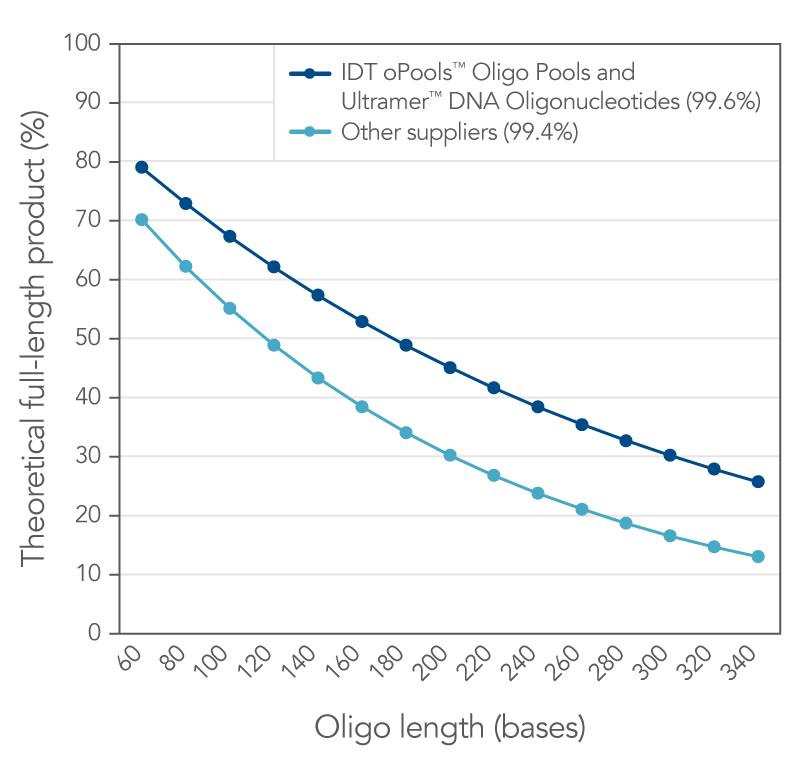
IDT proprietary DNA synthesis equipment permits rapid, high-quality synthesis of nucleic acids. This platform is adjusted to a lower throughput mode which uses an "extra rich" synthesis cycle for long oligos. Along with this refined synthesis cycle, Ultramer DNA Oligos use a solid support that is specifically designed to synthesize low-yield, high‑quality oligos up to 200 bases in length. Overall, these improvements to our already established manufacturing methods allow you to acquire longer, purer oligos for your research. Figure 1 illustrates the quality of Ultramer DNA Oligos.
Figure 1. IDT proprietary platforms have a better coupling efficiency than other suppliers, which provides more full-length oligonucleotides in your order. Small increases in coupling efficiency (≤1%) result in measurable increases in full-length product yield. The curves represent the theoretical yield for different lengths of oligos based on a coupling efficiency of 99.6% (IDT oligos, n = 34,627) and 99.4% (other supplier, n = 77 from three different suppliers) using the formula, percent full length product = (eff )(n-1)*100 where eff = coupling efficiency (for example, 99.4% = 0.994) and (n–1) is the number of coupling reactions needed to make an oligo of length n.
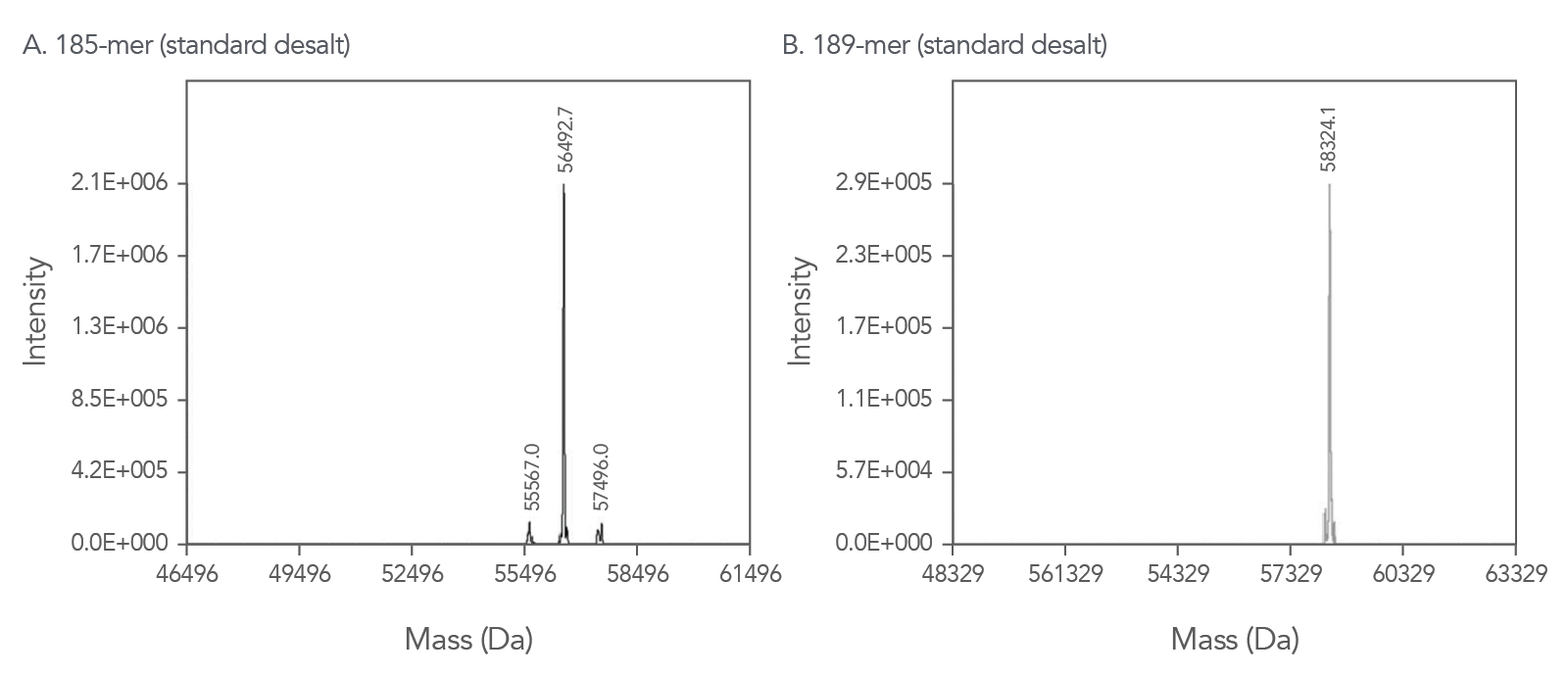
Our proprietary ESl-MS methods allow us to confidently provide quality control documentation for your oligos up to 200 bases in length*. These QC traces, such as the examples shown in Figure 2, are available online free of charge.
*With the exception of mixed base oligos, which could potentially represent multiple sequences and therefore cannot be evaluated by ESI-MS.
Services
Formulation
Diverse laboratory configurations and various individual preferences in the field require oligos that can be delivered in unique formats. With the formulation services listed below, you can have your oligos tailor-made to your specific requirements. Learn more »
Custom quality control
In addition to the free ESI-MS analysis that comes standard with your oligos,* your project may require one of the analytical services listed below. Learn more »
*With the exception of mixed base oligos, which could potentially represent multiple sequences and therefore cannot be evaluated by ESI‑MS.
GMP and OEM
If you require oligos that are ISO 13485:2016 certified, or if you are interested in our third-party manufacturing services, please click here.
Resources
Frequently asked questions
How do I convert my yield from nanomoles to ODs?
Calculating this conversion requires you to know the extinction coefficient for your sequence. With this, you can easily convert from your nanomole amount to ODs. To determine the extinction coefficient, you can analyze your sequence using IDT's free OligoAnalyzer™ Tool. Results from this analysis will also provide you with nmol/OD260 and µg/OD260 values.
Will you synthesize aptamers?
Yes. If you can provide the aptamer sequence(s), you can order directly from the IDT website.
To order online click on Custom DNA oligos, or if your sequence contains RNA bases, Custom RNA oligos.
Enter your desired scale, sequence, and purification.If you are ordering aptamers with a fixed sequence, IDT recommends HPLC or PAGE purification. For aptamer libraries containing random bases, IDT recommends standard desalt.
Will I lose more oligo when resuspending in polystyrene tubes compared with polypropolyene tubes?
All IDT oligonucleotides are shipped out in low absorption polypropylene tubes which can be used for long term storage without significant loss of material.
Why should I consider purification of my oligos?
Purification removes truncated products and other synthesis impurities. Our experience has shown that purifying an oligonucleotide that will be used in demanding applications saves both time and money in the long run. We recommend considering additional purification for any oligonucleotide that will be used for an application other than routine PCR, qPCR, or Sanger DNA sequencing.
As a general rule, IDT also recommends that any oligonucleotide longer than 40 bases should receive further purification. For questions, please contact us.
Why does the shipped quantity of product vary even for reorders of the same sequence?
Every IDT custom oligo is synthesized to order through a series of tightly controlled steps, including the coupling of the individual bases, cleaving the oligo from the solid support, desalting, and, if requested, purification. Each of these steps will result in a loss of final yield, which varies with each synthesis.
Due to this variation, IDT oligos are ordered according to the amount of starting material, also referred to as the scale. All custom oligos are assigned a minimum yield guarantee, but lot-to-lot variability in the final delivered quantity should be expected even when reordering the same sequence. For further questions about this subject, contact us.
Why do my primers have deleted 3' bases in the amplified fragment when I use a proofreading enzyme for PCR?
Proofreading polymerases (Pfu, Diamond Taq®, etc) have been shown to remove 3' bases from PCR primers [1].
If you experience this problem, you can protect your primer from degradation by adding one or more phosphorothioate bonds to the 3' end of your primer.
Also make sure to add the enzyme last, after the addition of dNTPs, in your PCR to minimize the risk of this issue taking place.
Reference
1. Skerra A. Phosphorothioate primers improve the amplification of DNA sequences by DNA polymerases with proofreading activity. Nucleic Acids Res. 1992;20(14):3551-3554.
Why are my oligos a yellow/brown color?
Customers who are new to working with large amounts of material are sometimes surprised when they see a yellow or brown discoloration to their oligos. If you have experienced this, do not worry. Color variation is normal in custom synthesis, especially as concentration increases. Importantly, IDT does not expect this to affect oligo function in our customers' experimental applications.

The image above depicts normal color variation as seen in oligo pools which are formulated to the same concentration (200 nmol oligos in 15 mL solution). In general, modified oligos at larger scales are more likely to display the variation seen above, while small-scale unmodified oligos (e.g., 25 nmol) tend to remain colorless in solution.
When will my oligos be shipped?
IDT oligos have various estimated ship dates depending on product type and composition. Oligos added to the shopping cart will show an estimated ship date for each line item.
Once the order is placed, this estimated ship date can be found on the order confirmation email as well as in the order history of your web account. You can find the order history at idtdna.com/site/orderstatus/orderstatus.
Select the order number you wish to review, and the status and estimated ship date of each line item will be shown.
IDT will email you a confirmation when your order has been shipped. This shipping confirmation will include the tracking number of the package, which you can use to monitor the status of your shipment. If you have additional questions about your order, please contact us.
When calculating the Tm of my oligo, should I only use the bases which are complementary to my target, or should I also include any sticky overhangs on the primer?
To determine the relative Tm of primers with non-complementary overhangs, only the complementary region should be taken into account.
You can obtain the Tm using the free, online OligoAnalyzer™ Tool.
What molar concentrations of oligonucleotide and salts are used to calculate the Tm reported on specification sheets?
The Tm value reported on our spec sheets uses a 25 µM oligonucleotide and a 50 mM salt concentration. It does not take into account dNTP or Mg2+ concentrations.
To get an accurate Tm for your specific application, all of these concentrations should be entered and adjusted to match the reaction conditions you plan to use.
The free IDT OligoAnalyzer™ calculates Tm for the concentrations you input.
What is the standard desalting procedure IDT uses for newly synthesized oligos, and what additional purification options do I have?
IDT uses a proprietary desalting technique that removes some truncation products and small organic contaminants from the synthesized oligonucleotide preparation. Removal of n-1mers produced during synthesis requires additional PAGE or HPLC purification.
PAGE is recommended for unmodified oligos >80−100 bases, while HPLC is the preferred method for oligos modified with either fluorescent dyes or attachment chemistries.
What is the stability of RNA vs. DNA?
RNA is inherently less stable than DNA due to its chemical structure. Additionally, RNases are more prevalent in standard laboratory conditions than DNases.
As even the slightest exposure to RNase can impact RNA stability, IDT has not performed rigorous long term stability studies for RNA.
What is the shelf life of my oligo?
The shelf life of an oligo is dependent on the temperature at which the oligo is stored and how the oligo is resuspended. Temperature is the more important of the 2 variables. Generally, oligos should be stored at –20°C. At this temperature an oligo has a minimum shelf life of 2 years, whether it is stored dry/lyophilized, in TE buffer, or in (non-DEPC treated) water.
Please see our DECODED™ article, Storing oligos: 7 things you should know, for data on oligonucleotide storage and a more thorough explanation.
What is the optimal length of a primer?
IDT recommends you aim for primers between 18–30 bases; however, the most important considerations for primer design should be their Tm value and sequence specificity. Primers should also be free of strong secondary structures and self-complementarity.
The free IDT OligoAnalyzer™ Tool can be used to evaluate these features.
What is the fidelity of your Ultramer™ oligonucleotide synthesis?
Standard DNA oligos have a coupling efficiency of 99.4%. Ultramer oligos have a 99.6% coupling efficiency. This increase results in higher fidelity synthesis. Regardless of synthesis choice, purification may be recommended for longer oligos based on your specific application.
For further information regarding Ultramer oligos, please see our Ultramer DNA Oligos catalog page.
What is the difference between scale and yield and why don’t I receive that full amount designated by the scale I order?
Oligo synthesis is accomplished through a series of steps, including coupling of individual bases, cleaving the oligo from the solid support, desalting, and if requested, purification of the oligo by HPLC or PAGE. No chemical reaction occurs with 100% efficiency, and, thus, each of these steps will incur a loss of final yield, which varies from specific sequence synthesis to synthesis.
Due to this variation, IDT custom oligos are ordered according to the amount of starting material used for the synthesis, referred to as the scale. While we cannot predict the actual final yield, we do guarantee a specific minimum yield for each oligo based on its sequence, starting scale, and the typical yield obtained under those specified conditions.
If you would like to know the minimum guaranteed yield for a specific oligo, simply add it to your Shopping Cart. The Shopping Cart provides you with the guaranteed minimum yield. If it is insufficient or excessive for your needs, simply edit the scale.
Please contact us if you have any questions about the yield you have received.
What is the best way to purify PCR products?
For most applications, it is best to purify PCR products by gel electrophoresis.
This simple method not only purifies the final PCR product but can be a valuable troubleshooting tool as well since nonspecific PCR products, primer-dimer products, negative amplification, and other elements can easily be identified by gel analysis.
What is CE service?
What does OD260 stand for?
The heterocyclic ring structures in DNA and RNA absorb light with a maximum absorbance near 260 nanometers (nm). An OD260, or optical density 260, is defined as the amount of light at a 260 nm wavelength which will be absorbed by an oligo resuspended in 1 mL water and the concentration is read in a 1 cm quartz cuvette.
This method of measurement is considered the most accurate means of assessing the amount of oligonucleotide present following synthesis. The relationship between measured OD260, molar extinction coefficient (ε260), and oligonucleotide concentration is given as: OD260 = ε260 x concentration.
What conditions should I use to HPLC purify my oligos?
IDT uses our own proprietary HPLC method to purify oligos in-house. A published set of conditions for an RP-HPLC protocol is:
- Hamilton PRP-1 Reverse Phase column 250 x 10 mm
- Buffer A = 100% ACN (acetonitrile)
- Buffer B = ACN/0.1 M TEAA pH 7.3
- Run at 4 mL/min over 30 min from 95–60% A
- Monitor at 260/297 nm
GMP refers to products manufactured under ISO 13485: 2016 QMS. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations for their legal marketing.