xGen™ HIV Amplicon Panel
A panel consisting of primers designed to cover 99.3% of the Human Immunodeficiency Virus (HIV) genome. These primers obtain high levels of genome coverage from as low as 1,000 double-stranded viral genome copies. xGen Amplicon Technology uses overlapping primer sets to provide comprehensive coverage in a single-tube, 2.5-hour workflow.
xGen NGS—made for HIV research.
Ordering
The IDT xGen HIV Amplicon Panel provides a single-tube, two-step PCR amplification workflow with primer sets designed to create amplicons across the entire HIV genome.
- >99% design coverage of the HIV genome
- Super amplicons result in increased coverage, even in changing or diverse genomes
- Sequence data from viral titers as low as 1,000 double-stranded viral genome copies
- cDNA to sequencer in 2.5 hrs
- Up to 1536 UDIs
Transform Your NGS Workflow with Automation
Looking to streamline your NGS workflows? Discover how automation can enhance efficiency and consistency in your lab with our NGS Automation solutions.
Request a consultation
Want to learn more about our xGen™ Amplicon Panels—developed with super amplicon technology—and how to customize your panel or spike-in genes in our predesigned panels? Your time is valuable—we’ll prioritize your inquiry and be in touch to discuss it ASAP.
Request a consultationProduct details
The xGen HIV Amplicon Panel offers a streamlined (cDNA-to-sequencer in 2.5 hours), single-tube NGS workflow for studying Human Immunodeficiency Virus (Figure 1). This Predesigned xGen Amplicon Panel built with xGen Amplicon technology includes amplicon tiling and creation of super amplicons to ensure comprehensive genome coverage across existing variant genomes and provide resistance to future viral mutations that may fall within a priming site, thus enabling future identification of novel variants (see our Tech Note on xGen Amplicon panels leverage super amplicon technology). Due to super amplicon formation, high level of coverage across the variant genomes was preserved as shown by % genome covered and % genome >10X coverage (Figure 2). xGen Amplicon Panel demonstrates high percent mapping and on-target reads to the HIV genome (Table 2). Identify the unidentified by using IDT’s solution for achieving comprehensive genome coverage of HIV.
Table 1. Features of the xGen HIV Amplicon Panel
| Features | Specifications |
|---|---|
| Design coverage and panel information | 107 amplicons, average amplicon size 102 bp Primers designed to target HIV (HXB2, GenBank K03455) Customize it here Minimum 1,000 double stranded viral genome copies |
| Input Material | 1st or 2nd strand cDNA, dsDNA |
| Time | 2.5 hours viral cDNA-to-library |
| Multiplexing capability for Illumina | Up to 1536 UDIs |
| Compatible with other indexes for Illumina? | Yes |
| Recommended depth for Illumina | 500,000 total reads per library, PE250 or PE150 |
Product data
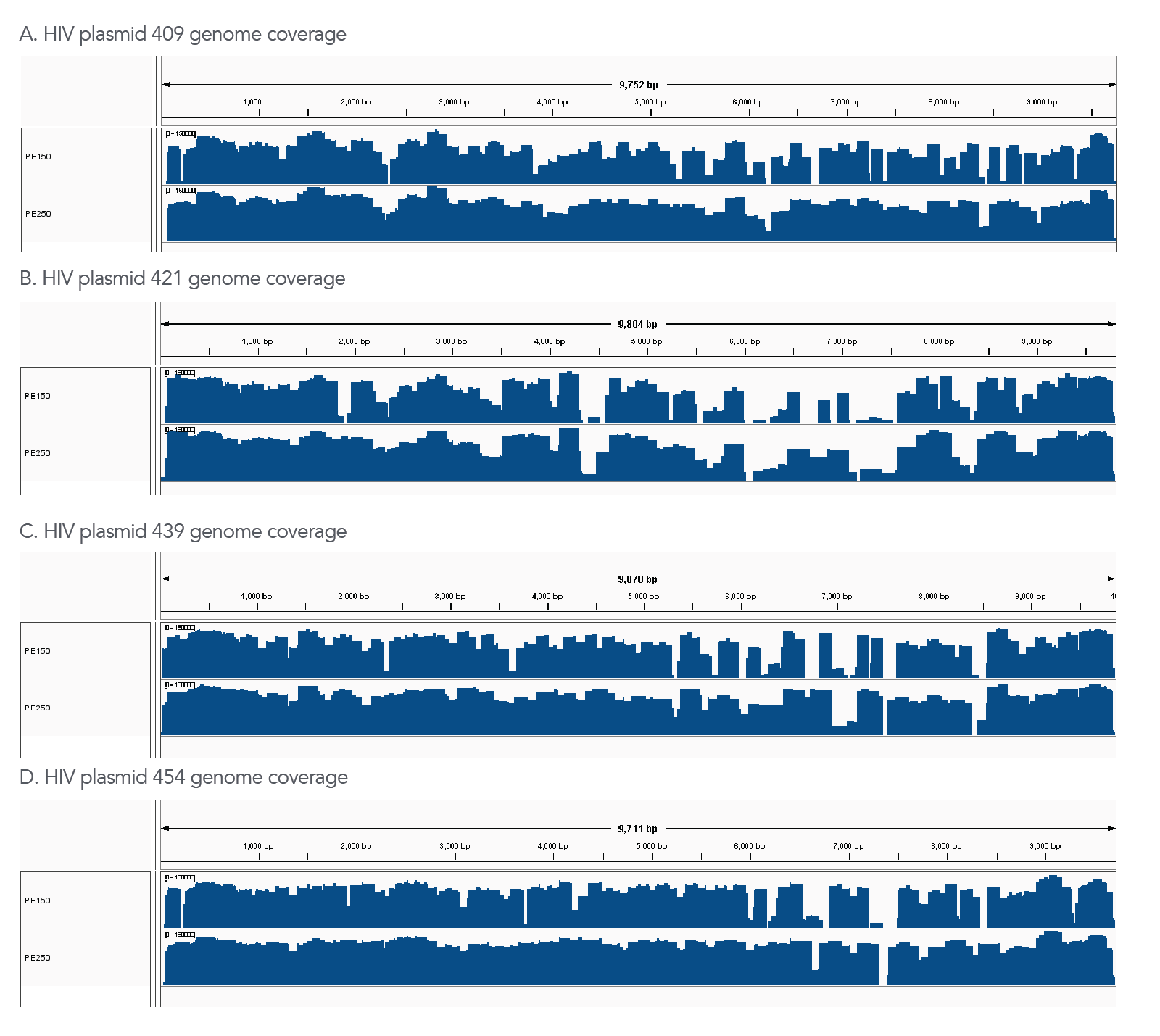
Figure 2. The xGen HIV Amplicon Panel provides a high level of coverage across variant HIV genomes. Examples of panel coverage are shown for four different HIV genomes. For all libraries, 1,000 plasmid copies carrying the complete HIV genome from four different strains ranging from 91–93% identity to the HXB2 reference genome were tested individually (plasmids described in Zaikos et al., 2018) in a background of 10 ng human gDNA (Coriell NA12878). The resulting NGS libraries generated with the xGen HIV Amplicon Panel were sequenced on a MiSeq™ (Illumina) with 150 bp and 250 bp paired-end sequencing (PE150, PE250). Resulting reads were downsampled to 500,000 total reads per sample for analysis. Example data for one replicate per HIV genome are shown in panels A-D (A) The input genome, plasmid 409, has 92% ID to the HXB2 panel design genome and data was aligned to the 409 reference. (B) The input genome, plasmid 421, has 91% ID to the HXB2 panel design genome and data was aligned to the 421 reference. (C) The input genome, plasmid 439, has 93% identity to the HXB2 primer design genome and data was aligned to the 439 reference. (D) The input genome, plasmid 454, has 93% identity to the HXB2 panel design genome, and data was aligned to the 454 reference. Despite the variation in these genomes to the HXB2 panel design genome, coverage is maintained across each genome due to super amplicon formation (see our Tech Note on how xGen Amplicon panels leverage super amplicon technology). The increase in coverage observed with the PE250 read length is due to improved coverage of the super amplicons spanning highly variable regions, which are not fully accessible with the shorter PE150 read length.
Table 2. xGen HIV Amplicon Panel NGS metrics on Illumina.*
| Plasmid 409 | Plasmid 421 | Plasmid 439 | Plasmid 454 | |||||
|---|---|---|---|---|---|---|---|---|
| PE150 | PE250 | PE150 | PE250 | PE150 | PE250 | PE150 | PE250 | |
| % identity to HXB2 design | 92% | 91% | 93% | 93% | ||||
| % mapping | 93.8% | 94% | 95.7% | 95.8% | 92.4% | 92.6% | 95.7% | 95.8% |
| % on-target (base) | 94.5% | 94.3% | 81.7% | 82.3% | 82.7% | 83.8% | 94% | 93.2% |
| % base uniformity | 53.9% | 66.6% | 53.9% | 57.5% | 65.4% | 76.4% | 75.9% | 87% |
| % genome >10x coverage | 92.5% | 99.3% | 80.9% | 91.5% | 88.5% | 98.6% | 91.7% | 98.5% |
| % genome covered | 97% | 99% | 92% | 98% | 95% | 99% | 96% | 100% |
Sequencing metrics are shown for the libraries described in Figure 2. Although the variant genomes had only 91-93% identity to the HXB2 reference genome used to design the panel, due to super amplicon formation, coverage across the variant genomes was preserved as shown by % genome covered and % genome >10X coverage. The panel was also specific in a background of human genomic DNA as demonstrated by high percent mapping and on-target reads to the HIV genome.
Resources
References
- Zaikos TD, Terry VH, Sebastian Kettinger NT, et al. Hematopoietic Stem and Progenitor Cells Are a Distinct HIV Reservoir that Contributes to Persistent Viremia in Suppressed Patients. Cell Rep. 2018;25(13):3759-3773.e9. doi:10.1016/j.celrep.2018.11.104