G-quadruplexes
In most contexts, when discussing DNA structure, we are talking about the genetic code and we think of the Watson and Crick model of DNA base pairing (Figure 1). However, other types of DNA base interactions regularly occur in the lab and in nature. One of the most common and important naturally occurring DNA structures is the G-quadruplex, which is perhaps most notably involved in the structure and function of telomeres.
What are quadruplexes?
G-quadruplexes are formed from the stacking of two or more G-tetrads (Figures 2−3). Tetrads form naturally in sequences with short G base repeats. Bases that are adjacent to tetrads form loop structures that help position a series of tetrads into a quadruplex stack structure (Figure 3). The intervening DNA loop bases can be any of the nucleotide bases, including G bases that are not involved in the tetrad structure. Tetrads also require the presence of a monovalent cation such as Na+ or K+ to balance the negative charge from the oxygen atoms at the center of the tetrad (Figure 2). The example shown in Figure 3 is one version of an intramolecular quadruplex, but up to four DNA strands can come together to make a quadruplex. Other DNA orientations are also possible, generating intramolecular varieties and making quadruplexes diverse structures that can be difficult to accurately predict [1].
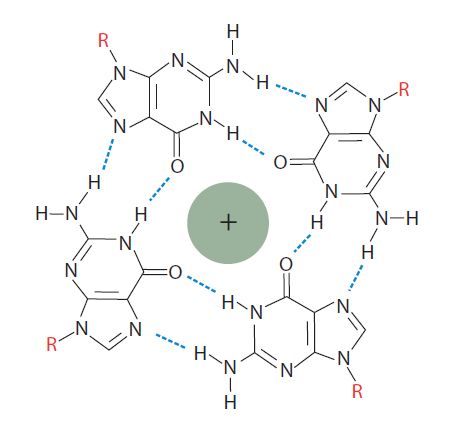
Figure 2. Guanosine tetraplex. In the presence of monovalent cations, closely spaced guanosines can interact with each other to generate a guanosine tetraplex. A tetraplex has 4 bases and can be formed from a single, looped nucleic molecule, or from as many as 4 separate RNA or DNA molecules that come together.
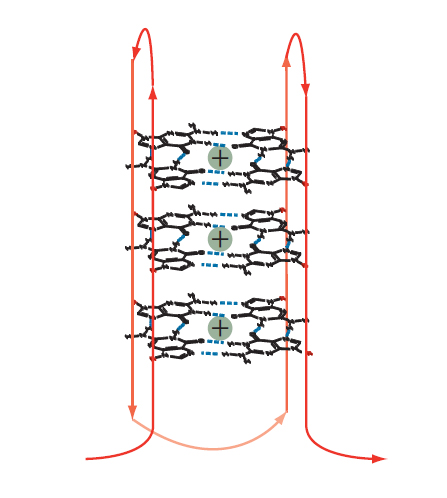
Figure 3. Intramolecular guanoisine quadruplex. Two or more tetrads can stack to form a complex secondary structure called a quadruplex. The example shown is formed from a single DNA strand (shown in red). Intervening bases are required to allow folding of the strand for positioning of the G bases such that they assume the individual, compact, tetraplex structures in the quadruplex stack.
Quadruplexes and tetrads in oligonucleotide synthesis
Multiple, intramolecular runs of G motifs 3−5 nt long and separated by short intervening sequences 1−7 nt long, have the best chance of forming quadruplexes. Oligonucleotides that form G-quadruplexes are more hydrophobic than Watson and Crick paired, double-stranded DNA, most likely because some of the polar groups are less available for aqueous interactions. This change in property makes these kinds of oligos more complex to manufacture and results in lower yields and purities. In the worst cases, yield can be reduced by more than half compared to a standard oligo synthesized at the same scale.
Quadruplexes also interfere with our ability to determine the purity of an oligo. Quadruplexes cause oligonucleotides to clump, producing multiple peaks on capillary electrophoresis. These additional peaks make it hard to quantify the amount of full-length product.
In some cases, to purify oligos with 4 or more consecutive Gs, or to measure the purity, RP-HPLC can relax the structure. For analyzing sequences with strong secondary structures (<–15 kcal/mol ΔG), it may be prudent to use analytical IE UPLC, pH 12, which usually denatures the sequence. However, these methods are not compatible with all modifications, and they do not solve structural issues in every sequence. Consequently, purity is not guaranteed for oligos with 4 or more consecutive guanine bases.
Quadruplexes and tetrads in oligo applications
For addressing secondary structure in most DNA and RNA, we recommend applications avoiding designs that have potential quadruplex or tetrad structures in either oligonucleotide or target sequences. Granted, this is not always possible.
G-quadruplexes can negatively affect experiments similar to other types of secondary structures, such as hairpin formation. While it may be difficult to predict how a given sequence will perform there are some guidelines that can help limit problems or troubleshoot issues arising from tetrad and quadruplex formation. The requirement for a monovalent cation to stabilize the tetrad and quadruplex structures underlines the importance of buffer conditions. Quadruplexes turn out to be more stable in the presence of potassium (K+) than sodium (Na+) ions. Therefore, when possible, adjusting monovalent cation concentration may improve experimental outcomes.
For PCR and qPCR applications, avoid placing primers and probes in or adjacent to regions within the amplicon where secondary structure of any kind is likely to occur, as this will affect PCR efficiency. This is also true for other applications involving hybridization, such as RNAi, where access to single-stranded RNA or DNA bases can be impaired by structure. Designing PCR and RNAi based methods to avoid G-repeat sequences, or selecting codons that limit G-repeats in protein expression applications are good ways to avoid potential problems.
It is important to remember that more G bases in either the oligo sequence or the target will increase the chances of tetraplex formation. These structures are usually not predicted by programs that look for secondary structures formed by standard Watson and Crick base pairing. However, if your sequence contains repetitive G motifs, and you are having problems with your experiments, quadruplexes are actually very common and may be the cause.


