SYBR® Green dye and PrimeTime™ qPCR Probe Assays
Overview
qPCR for gene expression analysis can be done with primers and intercalating dyes such as SYBR® Green (Thermo Fisher Scientific) or with primers and fluorescent probes such as PrimeTime™ qPCR probes. Understanding the key benefits of each method will help you design a high-quality assay for your research.
Basics of quantitative PCR (qPCR)
The polymerase chain reaction (PCR) amplifies a target sequence from a sample of DNA or RNA (that has been converted into cDNA). In quantitative PCR (qPCR) also known as real-time PCR the amount of DNA product is measured in each amplification cycle based on a fluorescent signal. The fluorescence is produced using intercalating dyes, such as SYBR® Green dye (Thermo Fisher Scientific) or using 5’ nuclease probes such as PrimeTime™ qPCR Probes. The accumulation of a PCR amplicon in each cycle is detected by graphically plotting the cycle number on the x axis and the fluorescence on the y-axis. Most commonly used are copy number standards run alongside the unknowns to generate a standard curve. By comparing the Cq value of the unknown to the standard curve values, the amount of starting DNA can be determined. qPCR reactions can also be coupled with reverse transcriptase (RT) to quantify the abundance of an RNA transcript, which is sometimes referred to as RT-qPCR, qRT- PCR, or real-time RT-PCR. These assays are used to assess gene expression─ a key measure for understanding biological systems including development and splice variant-specific gene expression. Real-time PCR can also be used in genotyping applications, such as identifying copy number variations, determining frequency of SNPs in populations, multi-species variations, or association studies to determine genes relevant to specific traits.
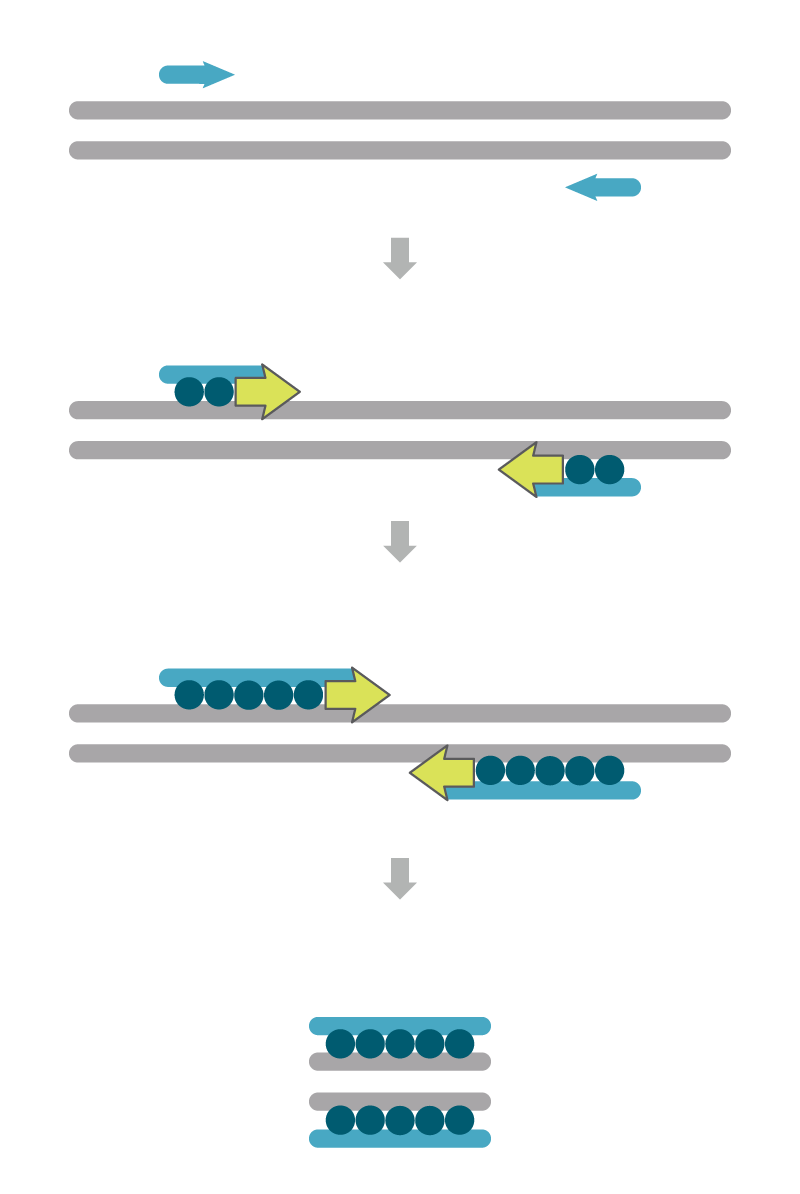
Intercalating dye or primer assays
The number of amplicons created during PCR amplification can be measured using intercalating dyes such as SYBR® Green (Thermo Fisher Scientific), SYTO® (Life Technologies), EvaGreen® (Biotium), and LCGreen® (Idaho Technology, Inc.). These are sometimes called primer-only assays (Figure 1). These dyes do not fluoresce until they bind to double-stranded DNA. The strength of the fluorescence signal is proportional to the relative amount of PCR amplicons present after extension.
Although intercalating dyes are inexpensive, there can be drawbacks to their use. If the reaction has issues with mispriming of non-specific sites, cross- reacting genes, or the accumulation of primer-dimer products, intercalating dyes will bind to the incorrect double-stranded DNA along with any actual desired PCR amplicon. As such, it is important to confirm that the PCR reaction produces only one single PCR product, which can be done by melt curve analysis and gel electrophoresis. The dyes are also non-specific which renders multiplex qPCR assays, a key for genotyping applications, impossible with this approach.
For more information on melt curve analysis, see the DECODED™ online article, Explaining multiple peaks in qPCR melt curve analysis. Another important resource for understanding whether your qPCR amplicon includes non-specific sequences is the free online uMeltSM software (University of Utah) that predicts melt curves and their derivatives.
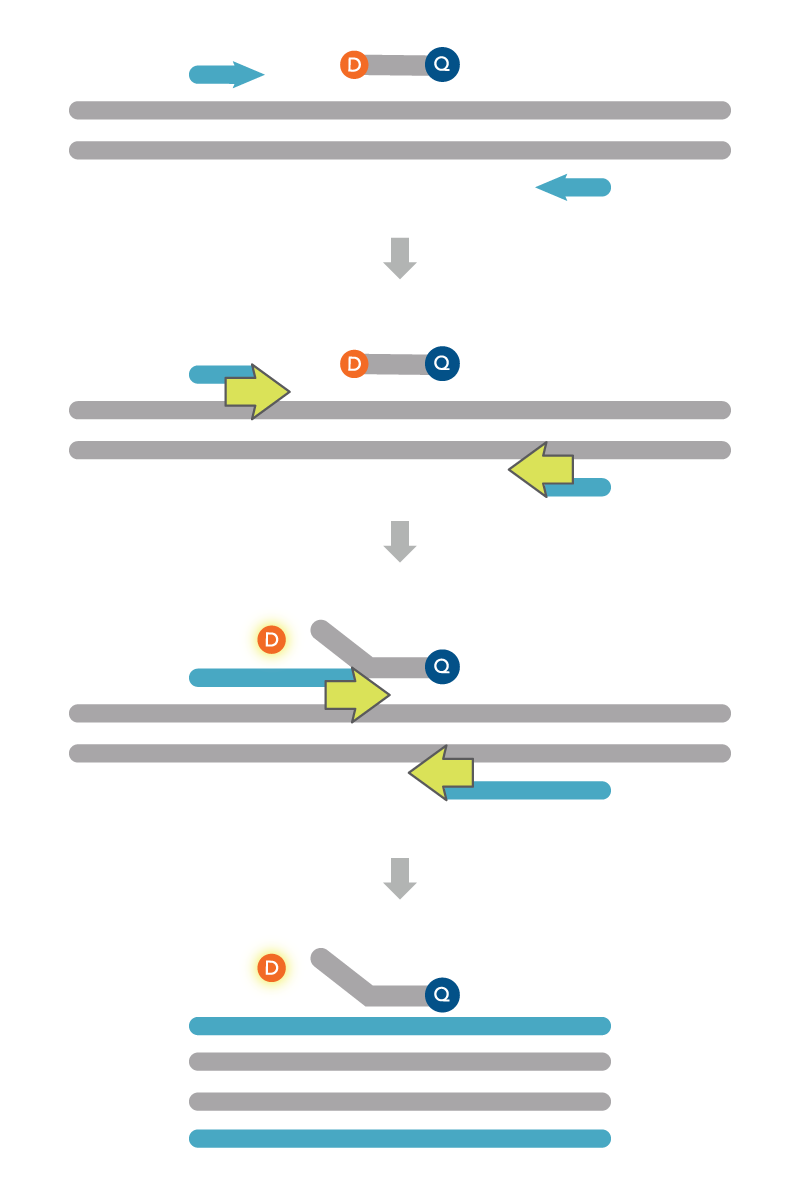
5’ nuclease or probe-based assays
Probe-based, or 5’ nuclease assays, enable the quantification of an accumulating, discrete PCR product (Figure 2). Also known as PrimeTime or TaqMan® assays (Thermo Fisher Scientific), these assays include a fluorescently labeled oligonucleotide probe in addition to the forward and reverse PCR primers. The combination of probe and primers increases the precision in the amplification reaction since all three must bind to the correct target for a fluorescence signal to be generated. Probe-based assays also lets users combine multiple primer and probe sets in one reaction. These multiplex qPCR assays include unique 5′ fluorophores for each of the probes, allowing the wavelength of fluorescence signal to indicate the presence and relative abundance of the transcript. The ability to identify these signals in any probe-based assay depends on having the instruments and software capable of recognizing the different fluorophores. For more information on selecting the different dyes for multiplex assays, see Recommended dye combinations for multiplex qPCR. IDT also has a Multiplex Dye Selection Tool that offers guidance on the conventional and alternative fluorophores to use with many qPCR instruments.
PrimeTime qPCR Probes are 5' nuclease probes, with a 5′ fluorophore and a 3′ quencher. The probe hybridizes between the two PCR primers. Since the quencher and fluorophore are in close proximity within the same fragment, no fluorescence is recorded. During annealing, the probe and PCR primers attach to their complementary sequences in the target region. The probe is designed with a higher melting temperature and will bind first. During extension/elongation, the 5’ to 3’ exonuclease activity of Taq DNA polymerase hydrolyzes the probe. Once hydrolyzed, the 5’ fluorophore is released from the quencher. Without a quencher nearby, the fluorophore emits a signal that the qPCR instrument can record. The amount of fluorescence, in turn, correlates with the amount of PCR products that are synthesized.
It is important that probe length is short, so the quencher is close enough to absorb the fluorescent energy. Longer probes will have high background fluorescence since the quencher will be less effective due to the separation distance. For more information on strategies to design qPCR probes with low background, see How to design primers and probes for qPCR and PCR, Double-quenched probes increase qPCR precision, and Improve assays with customizable oligos and probes containing cost-effective locked nucleic acids.
Converting primer-only assays to probe-based assays
SYBR Green and other types of intercalating dyes are more cost-effective for the researcher than 5′ nuclease probe assays, but can pose issues if the primers form primer-dimers, bind to non-specific sites, or anneal to cross-reacting genes. PrimeTime qPCR Primer Assays include predesigned assays for human, mouse, or rat targets. These assays have been designed with advanced bioinformatics to avoid these issues. The primer pairs are designed and premixed for real-time PCR, which makes the reaction setup easy. The primer sequences are also identical to a coordinating PrimeTime qPCR Probe Assays. This means you can easily transition from primer assays to 5′ nuclease assays to better target nucleic acid fragments or establish a new multiplex assay.Real-time qPCR guide: Design, analysis, and troubleshooting
Familiarize yourself with critical parameters of qPCR assay design. This complete 62-page guide covers every aspect of probe-based qPCR assays.
qPCR applications
Gene expression
Gene expression is the conversion of heritable, genetic information into RNA or protein. Differences and changes in gene expression are important measures for understanding biological systems, including normal development and disease progression.
Genotyping
Genotyping is the process of determining the DNA sequence, called a genotype, at specific positions within the genome of an individual. Sequence variations can be used as markers in linkage and association studies to determine genes relevant to specific traits or disease.
Genotyping by digital PCR (dPCR)
Digital PCR (dPCR) includes the same reagents found in a typical qPCR assay and amplified in a similar manner, but dPCR divides the reaction into nano-sized droplets or wells before amplification. The partitions are so small that either 1 or 0 templates are in each. After amplification, the fluorescence in the well or droplet represents the genotype.
Products for qPCR
PrimeTime qPCR Probe and Primer Assays
PrimeTime qPCR Probe Assays are 5′ nuclease assays that include a library of predesigned sequences for human, mouse, and rat targets. Custom assays may be created for these or other species using the PrimerQuest Tool. Predesigned sequences and custom assays are available in tubes or plates.
PrimeTime qPCR Primer Assays for use with intercalating dyes, such as SYBR® Green dyes, include predesigned sequences for human, mouse, or rat targets. Custom assays may be created for these and other species using the PrimerQuest Tool. Predesigned sequences and custom assays are available in tubes or plates.
Affinity Plus™ qPCR Probes
Use Affinity Plus qPCR Probes for enhanced discrimination of thermodynamically similar samples such as single nucleotide polymorphisms and transcript variants. The Affinity Plus bases used in these qPCR probes include up to 6 locked nucleic acid monomers. When incorporated into a probe, locked nucleic acids impart heightened structural stability, leading to increased hybridization melt temperature (Tm).
xGen NGS Amplicon Sequencing
Our amplicon solutions enable you to advance from sample to sequencing faster, without sacrificing coverage or yields. If your research requires rapid, ready-to-sequence materials or you are working with low-input quantities, xGen NGS can unlock the answers you seek.
RUO23-1664_001