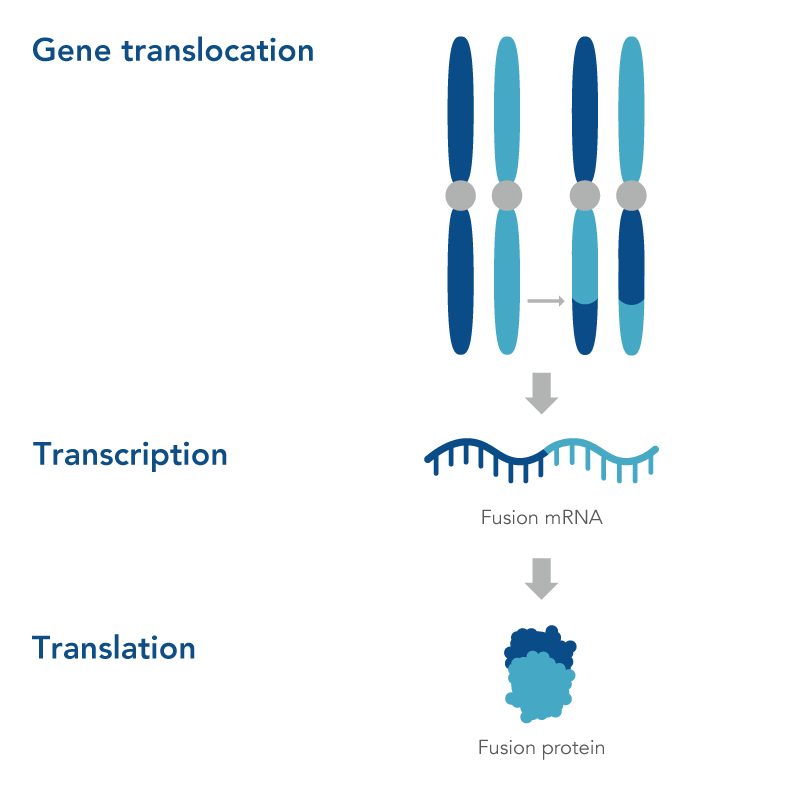
What is gene fusion?
Gene fusion occurs when two genes are ”fused” or joined, forming one new fusion gene that typically has an altered phenotypic function. This can happen either when there’s an error in the gene transcription process, in which genes are transcribed into RNA and then translated into proteins, or during a genomic structural rearrangement (e.g., translocation, inversions, deletions/insertions) that goes awry (Figure 1)[1].
Why are gene fusions important?
Fusion genes are key biomarkers in cancer research as the expression of these genes can lead to the formation of fusion proteins with oncogenic properties, abnormal expression of proto-oncogenic genes, or the loss of expression of tumor-suppressing genes [1]. Knowledge of fusion genes that are present in a sample can provide researchers with a better understanding of how certain types of cancer progress as well as provide important information for genomic medicine researchers [2].
Are gene rearrangements always associated with cancer?
In humans, regulated gene rearrangements can be beneficial, such as V(D)J recombination for adaptive immunity. When occurring elsewhere in the genome however, they are often harmful drivers of cancer. In other organisms (e.g., bacteria and plants) gene fusions have been suggested to be an important evolutionary mechanism [3].
The first human fusion gene was discovered in the 1960s [4], and since then over 10,000 fusion genes have been identified and determined to be potential cancer research biomarkers [5]. The expression of fusion genes can result in fusion proteins and abnormal gene expression. This change in expression or generation of fusion proteins have been shown to contribute to the formation of tumors in some cancer types [1].
How are gene fusions detected
Gene fusions can be identified through a number of approaches including:
- Fluorescence in situ hybridization (FISH)
- Reverse transcriptase PCR (RT-PCR)
- Microarrays
- Whole genome sequencing (WGS)
- Targeted hybridization capture
- Amplicon sequencing
- RNA sequencing (RNA-seq)[5]
- Anchored Multiplex PCR (AMP™) Chemistry
Molecular approaches such as FISH and RT-PCR are relatively fast and affordable. These approaches are limited in that they only target one fusion gene at a time and knowledge of the fusion gene’s sequence is required prior to detection. Next generation sequencing (NGS) techniques like WGS, RNA-seq, and AMP Chemistry are useful in that they aren’t limited to known targets and allow users to multiplex many targets, generating a large amount of data in a single sequencing run [5].
When NGS approaches are used to detect gene fusions, bioinformatic analyses are also needed. The basic pipeline for pre-processing sequencing reads generally consists of quality control steps (i.e., removing low-quality reads), alignment of reads, and duplicate filtering. Once reads have been pre-processed a gene fusion detection software can be used [5].
Visualization of gene fusions with Archer™ Analysis
Detecting and visualizing fusion breakpoints (areas in a genome where gene fusions may have occurred) are important in cancer research. This can be accomplished using Archer Analysis which includes a user-friendly bioinformatics platform interface, designed for identification of gene fusions, splice variants, and other important cancer research biomarkers. Archer Analysis is designed to be used with all Archer Research Assays that are powered by AMP Chemistry to detect key cancer biomarkers, like gene fusions.
To find out more about Archer Research Assays and Archer Analysis click here.