Need a quick way to find primers within a gene or the expected size of the resultant PCR product? Here we show you how to get this information using NCBI's BLAST.
NCBI’s BLAST (Basic Local Alignment Search Tool)
NCBI’s BLAST (Basic Local Alignment Search Tool) is an incredibly powerful tool that efficiently queries the massive Genbank® database. However, due to the heuristic nature of NCBI BLAST and removal of low complexity data, queries for short sequences like primers often return incomplete data.
Use these tips to refine Primer-BLAST results:
- Concatenate the two primer sequences into one sequence separated by 5–10 Ns and enter into BLAST sequence box.
- Before submitting, narrow the search by selecting the species, if known; otherwise, choose Nucleotide Collection (nr/nt). If you’re looking for RT-PCR primers, select the reference mRNA sequences (refseq_mRNA) database.
- Under Program Selection, select the Somewhat similar sequences (blastn) program.
- Under Algorithm parameters, decrease word size to 7, increase expect threshold to 1000, and turn off the low complexity filter.
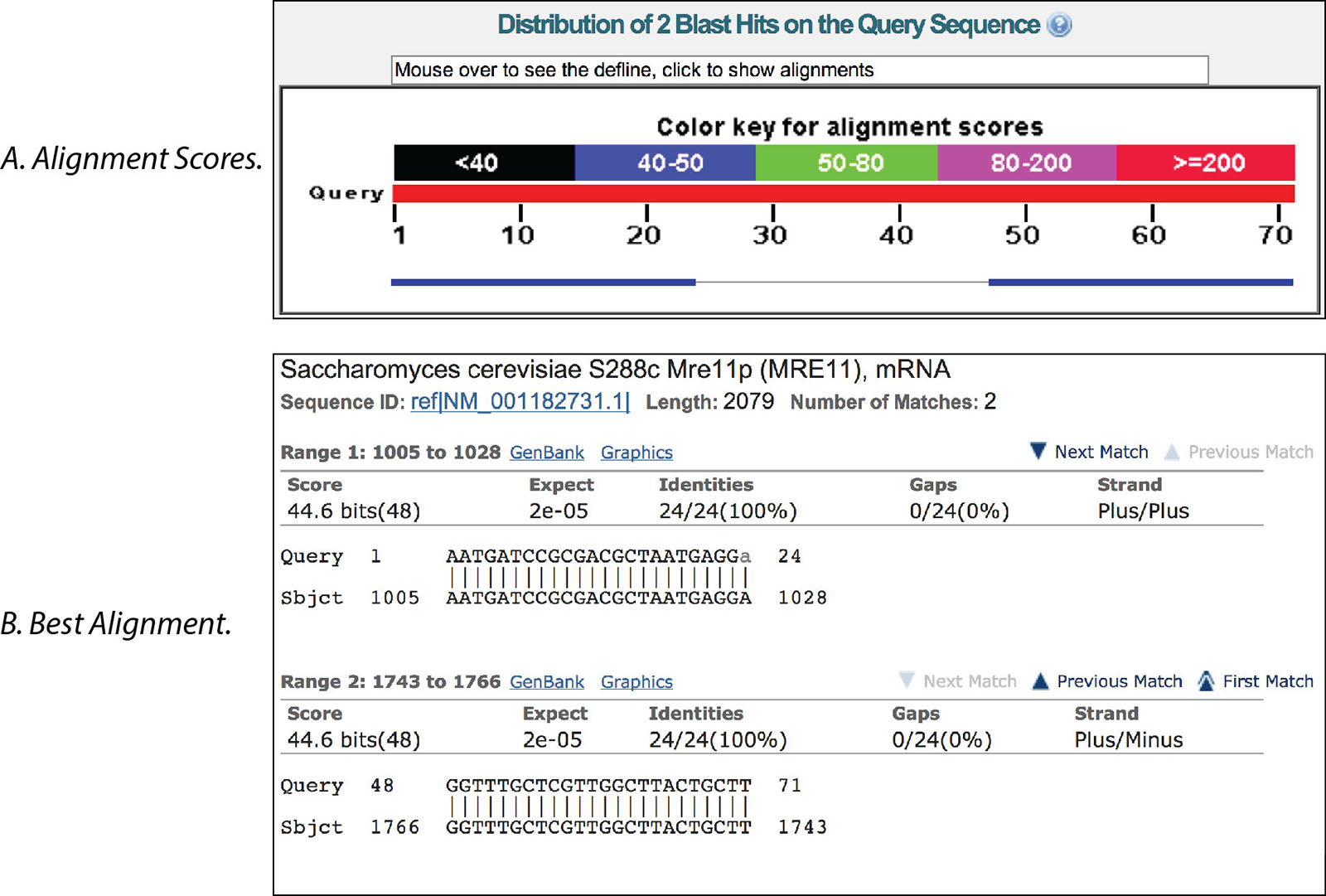
In the example below, the results with a line connecting the 2 boxes indicate the 2 primers are in the same sequence (Figure 1A). Clicking on these results shows location within the sequence (between bases 1005−1766 of the MRE11 transcript in yeast) and indicate the expected PCR product should be 762 bp in length (Figure 1B).
Want more information about your oligos? Check out IDT’s OligoAnalyzer™ Tool.
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations. Doc ID: RUO23-1933_001

