Background
Scientists merge new and old methods of genome visualization with the power of CRISPR to show chromosomal organization within the cell at a greater depth and closer scale than previously realized.
FISH technique
The scientific community has known for more than a few decades that the spatial DNA organization within eukaryotic cells is critical to its overall genome function, including gene expression [1]. To delve deeper into these phenomena, researchers working in the field of microscopy began using a technique called “fluorescent in situ hybridization” (or FISH for short), looking for chromosomal abnormalities—deletions and duplications—that can cause disease. How FISH works is by hybridizing oligos and attaching fluorescent molecules to probes that bind to complementary regions of DNA, revealing the precise location of that DNA and providing invaluable information about the structure and organization of the nucleus.
FISH and related in situ hybridization methods continue to provide the necessary data for mapping the positions of individual genes on chromosomes, but they are by no means perfect methods. Permeabilizing cells and denaturing DNA so that the oligos can access and bind to genomic DNA is tricky business. Insufficient permeabilization or denaturation leads to low or no signal, while too much can result in a destruction of the delicate features and structure of the nucleus. Even in the best case, some loss of structure usually occurs with FISH.
Chromosomal-scale visualization with Oligopaints
Researchers from Dr Ting Wu’s lab (Harvard), including Jeffrey Moffit (Harvard), Brian Beliveau (Univ. of Washington) and Eric Joyce (Univ. of Pennsylvania) codeveloped a technology for visualizing chromosomes in situ to reveal the greatest level of organizational detail so far in a 3D genome, evolving into what has become known as Oligopaints. Essentially, Oligopaints is a flexible, oligo-based technique that uses FISH to paint details, from single copy regions to megabases [2]. This tool has become increasingly powerful since it takes DNA from flow-sorted or micro-dissected chromosomes as FISH probes [3].
Chromosomal-scale visualization with GOLD FISH
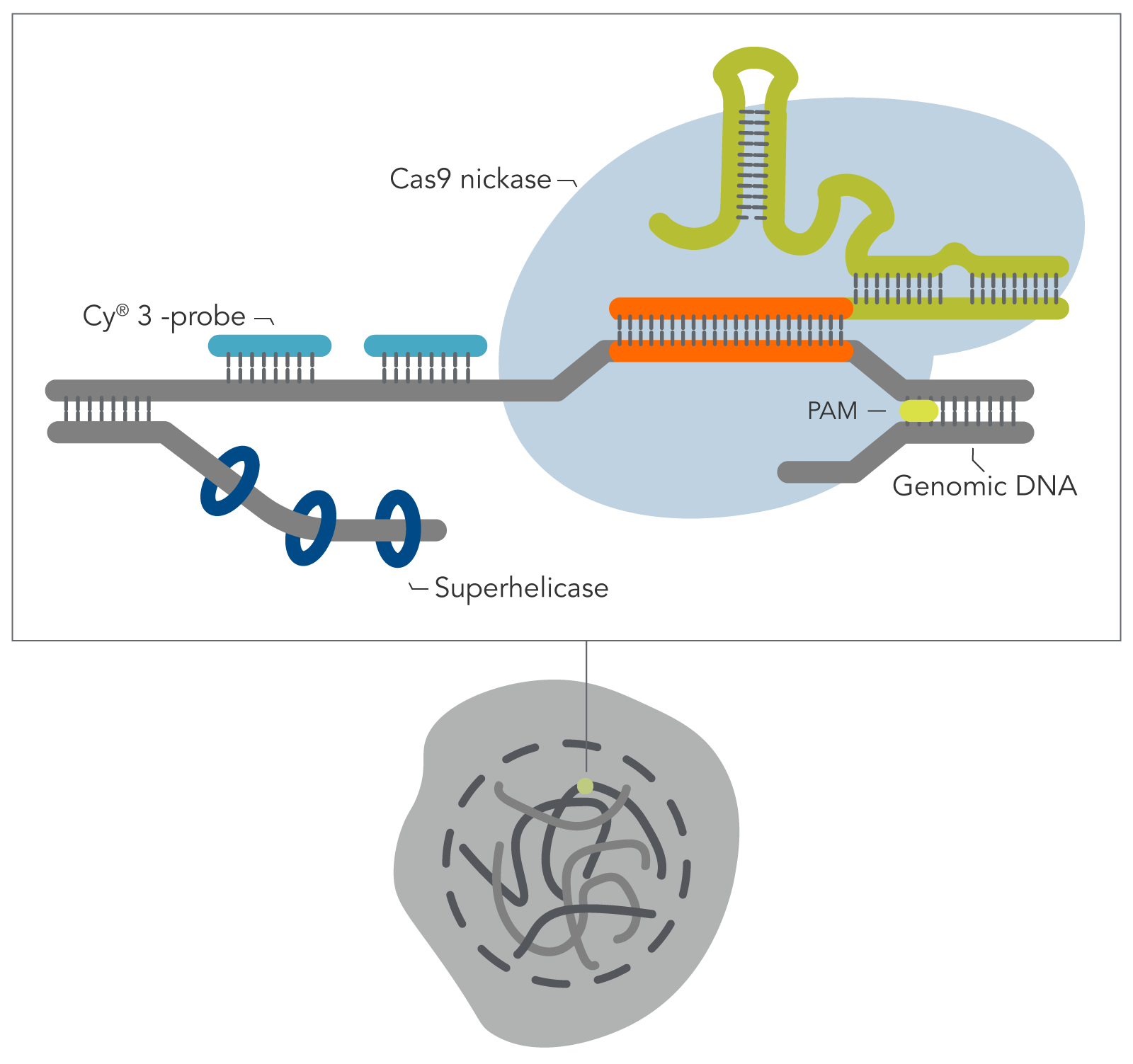
Building on the work from the Wu lab by combining the visualization breakthroughs of Oligopaints, with the ability of a CRISPR Cas9 enzyme to access DNA that is often tightly bound and inaccessible in nucleosomes, Yanbo Wang and associates (John Hopkins) have developed a new DNA FISH method—genome oligopaint via local denaturization (GOLD) FISH. With the GOLD FISH innovation, a Cas9-helicase fusion protein targets and denatures gDNA for probe binding. In contrast to FISH, GOLD FISH takes this modified Cas9 protein that targets superhelicase activity to specific genomic regions, and locally unwinds the DNA at physiological temperatures.
Experiment
In the Molecular Cell article featured here, GOLD FISH was performed on different genomic sequences, i.e., repetitive, non-repetitive, and whole chromosome ‘paint.’ Individual GOLD FISH experiments were performed with different parameters such as Cas9 RNP and oligo* probe concentrations. Detailed protocols of each GOLD FISH experiment presented in this work can be found in Methods S1 of the article.
Results and discussion
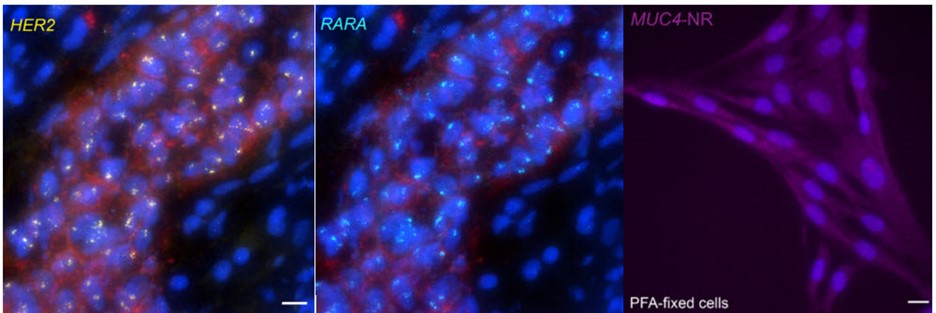
When contrasting this newer technique over conventional DNA FISH, instead of global denaturization by heat or chemicals, GOLD FISH locally denatures targeted chromatin in a much milder way by loading a superhelicase at a Cas9-generated nick. This targeted chromatin denaturing method both minimizes nonspecific binding of probes and provides superb signal and specificity. Those qualities in combination enable a richer understanding of nonrepetitive gDNA loci, highlighting a bright contrast between active and inactive X chromosomes [4]. As realized during these experiments, GOLD FISH can be used for rapid identification of HER2 gene amplification in patient tissue. Understanding that GOLD FISH is scalable—from a single locus to chromosomal-scale “paint”—makes it highly desirable to study pathologic tissue samples.