Types of DNA and RNA Sequencing

Sequencing is an integral part of genetic research and discovery. It’s also evolving at a rapid pace, so we’ve compiled this helpful list, which outlines the different types of DNA and RNA sequencing, as well as epigenomics.
If you find this list of types of DNA and RNA Sequencing helpful, you might want to download our free Next Generation Sequencing Guide, which includes this information, the recent history of NGS, the steps in sequencing, and much more.
Types of DNA and RNA Sequencing
Types of DNA Sequencing
1. Whole genome sequencing (WGS)
Whole genome sequencing is used to initially determine the genome sequence of an organism. Additionally, WGS can be used to determine variant (mutation) frequencies within populations of organisms and to associate genetic variants with disease through genome-wide association studies (GWAS). A GWAS performs WGS on two populations and compares trait differences with genetic differences to associate identified traits with identified variants. WGS was first used for clinical diagnostics in 2009, but time and costs have limited its use in this area [1,2]. As the price of WGS decreases, it is becoming more common as a diagnostic tool. Having achieved the “$1000 genome” [4], multiple companies are pushing towards the goal of the “$100 genome” [5,6].
2. Targeted sequencing
Targeted NGS allows users to sequence specific areas of the genome for in-depth analyses in a more rapid, cost-effective way than whole genome sequencing. Targeted sequencing detects known and novel variants within your region of interest. This method generally produces a smaller amount of data than WGS, making analysis more manageable.
There are several methods of targeted sequencing, each appropriate for specific applications. The most popular methods are hybridization capture, amplicon sequencing, and molecular inversion probes (MIPs). For more in-depth comparison of hybridization capture and amplicon sequencing, see our Targeted Sequencing Guide.
3. Whole exome sequencing (WES)
Whole exome sequencing identifies all the protein-coding genes in the genome. Focusing on protein-coding exons (and excluding other regions of the genome) can lower the cost and time of sequencing, as exons make up only 1% of the genome. Variants in protein-coding exons are responsible for many diseases, so this level of sequencing is often sufficient for diagnostic applications. WES is a more practical method for mapping variants that are rare in the population to elucidate complex disorders [7]. It is also a feasible option for discovery science [8]. WES is particularly useful in oncology research and is currently used for cancer diagnostics [9]. Information gained from WES can provide insight into prognoses and personalized treatment options [10]. WES is most often carried out with hybridization probes rather than amplicons.
4. Hybridization capture
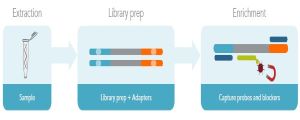
Prior to hybridization capture, samples are converted into sequencing libraries. Regions of interest in this library are then captured using long oligonucleotide biotinylated baits (Fig. 1). Because the DNA was random sheared during library preparation, captured fragments are overlapping and unique. Baits can be tiled, overlapped, and positioned to overcome challenges of repetitive sequences, etc. With advanced design, capture can be made very uniform. Hybridization capture is often used for targeted exome sequencing. Other applications include genotyping, rare variant detection, and oncology diagnostics [11].
Figure 1. Hybridization capture workflow
5. Amplicon sequencing
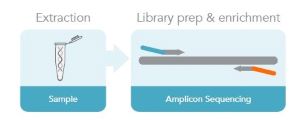
Amplicon sequencing is a highly targeted approach that enables you to analyze variation in specific genomic regions. This method uses PCR to amplify DNA to make amplicons. The amplicons are indexed and sequenced (Fig. 2). Amplicon sequencing is typically used for disease-associated variant detection and diagnostics [12]. It can also be used for genotyping by sequencing and to confirm CRISPR genome edits. Read more about how you can evaluate CRISPR-Cas9 edits quickly and accurately with rhAmpSeq targeted sequencing.
Figure 2. Amplicon sequencing workflow
6. Molecular inversion probes (MIPs)
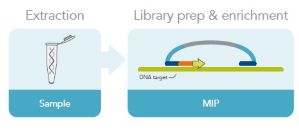
Molecular inversion probes are another common target enrichment method. Targeted-specific sequences are ligated to both ends of a universal sequence to make the MIP. The MIP hybridizes to the region of interest before a gap-filling reaction and a second ligation closes the circles. A restriction enzyme in the MIP may be used to create a linear molecule. The target sequences are amplified before sequencing (Fig. 3). MIPs are particularly useful for large-scale genotyping.
Figure 3. Molecular inversion probe workflow
Types of RNA Sequencing
1. Whole transcriptome sequencing (WTS)
Since RNA expression level can vary based on cell type and disease state, understanding the transcriptome can provide valuable insight. Sequencing the whole transcriptome provides the most comprehensive data because it contains information regarding both coding and noncoding RNA and both known and novel variants. A variation of Whole Transcriptome Sequencing, or RNA-seq, is stranded RNA-seq, which retains information about the strand of origin of transcripts. This makes it possible to identify novel transcripts, including antisense RNAs. RNA-seq is often used for discovery science.
Typically, RNA-seq is used to evaluate messenger RNA (mRNA) expression levels and can also be used to evaluate changes in gene expression over time, gene fusions, single nucleotide polymorphisms (SNPs), alternative splicing, and RNA modifications.
2. Targeted gene expression with RNA-Sequencing
Like DNA, RNA can be targeted for sequencing using hybridization capture or amplicon sequencing technologies. Specific populations of RNA can be targeted. Most often, coding transcripts are targeted, but other populations of RNA such as tRNA (transfer RNA) or small RNA may be enriched.
3. Ribosomal RNA depletion
Ribosomal RNA (rRNA) makes up 80-95% of the cell’s RNA; however, since its expression is constant, it is rarely of interest. Therefore, it can be advantageous to avoid sequencing these molecules and focus your sequencing on more useful data. rRNA may be removed from samples by 1 of 2 methods: 1). Biotinylated probes can be used to bind the rRNA and thus remove it from the RNA sample. 2). DNA probes that bind the rRNA can be used in conjunction with RNase H to degrade the rRNA from the sample library before library prep.
Epigenomics
1. ChIP-Sequencing
Chromatin immunoprecipitation (ChIP) is used to evaluate protein interactions with DNA. Protein can regulate DNA, impacting its expression. This type of regulation can be influenced by the environment and can change over time. ChIP identifies sites in the DNA sequence where protein is bound. An antibody is used to bind the protein of interest, which allows immunoprecipitation of the DNA bound to the protein.
When this type of identification is done using an array, it is called ChIP-chip. Evaluating protein interactions of the whole genome by sequencing is called ChIP-Seq. ChIP-Seq experiments usually focus on transcription factors, histones, and histone modifications so they can reveal information about gene regulation, cell proliferation, and disease progression.
2. Methyl-seq
Methylation, another method by which DNA is regulated, is also influenced by the environment and can change over time. Methyl groups are added to the DNA sequence and can repress DNA expression.
Methyl-Seq, also known as bisulfite sequencing, treats the DNA with bisulfite before sequencing to provide information about the methylation status of the DNA sequence. This information is primarily used to evaluate gene-environment interactions.
We hope this information helps your sequencing. For more information, download our free Next Generation Sequencing Guide.
REFERENCES
- Rieber N, Zapatka M, et al. (2013), Coverage bias and sensitivity of variant calling for four whole-genome sequencing technologies. PLos One 8(6):e66621.
- Welch JS, Westervelt P, et al. (2011). Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. Jama 305(15): 1577-1584.
- Lunshof JE, Bobe J, et al. (2010). Personal genomes in progress: from the human genome project to the personal genome project. Dialogues Clin Neurosci 12(1):47-60.
- Hayden EC (2014). Technology: The $1,000 genome. Nature 507(7492):294-295.
- Herper M (2017). Illumina promises to sequence human genome for $100 – but not quite yet. Forbes www.forbes.com/sites/matthewherper/2017/01/09/illumina-promises-to-sequence-human-genome-for-100-but-not-quite-yet. Accessed November 5, 2019.
- McMorrow D (2010). The $100 genome: Implications for the dod, MITRE CORP MCLEAN VA JASON PROGRAM OFFICE.
- Williams HJ, Hurst JR, et al. (2016). The use of whole-exome sequencing to disentangle complex phenotypes. Eur J Hum Genet 24(2):298-301.
- Bamshad MJ, Ng SB, et al. (2011). Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 12(11):745-755.
- Kamps R, Brandao RD, et al. (2017). Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci 18(2):308.
- Rabbani B, Nakaoka H, et al. (2016). Next generation sequencing: implications in personalized medicine and pharmacogenomics. Mol Biosyst 12(6):1818-1830.
- Necchi A, Bratslavsky G, et al. (2019). Genomic features for therapeutic insights of chemotherapy-resistant primary mediastinal nonseminomatous germ cell tumors and comparison with gonadal counterpart. Oncologist 24(4):e142-e145.
- Shin S, Kim Y, et al. (2017). Validation and optimization of the Ion Torrent S5 XL sequencer and Oncomine workflow for BRCA1 and BRCA2 genetic testing. Oncotarget 8(21):34858-34866.